Advanced Molecular & Cell Biology 1
DNA Structure; RNA Polymerase Properties; $\sigma$ Factor 70
!!! Main purpose for this lecture
- DNA sequences drive specific Protein:DNA interactions, which is fundamental for processes like transcription.
- Transcription steps can be experimentally defined, allowing for detailed understanding and manipulation.
General Properties
- Response Element Recognition: Response elements are DNA sequences that are recognized and bound by transcription factors, enabling regulation of gene expression.
- HAT/PTM Complexes: Histone acetyltransferases (HAT) and post-translational modifications (PTM) play pivotal roles in regulating gene expression by modifying histones and other proteins.
- Chromatin Remodeling: This process allows for accessibility of transcriptional machinery to the DNA by changing the structure of chromatin.
- Mediator: A complex that acts as an interface between transcriptional activators, RNA polymerase, and other transcriptional machinery.
The environment inside a cell is overcrowded, causing molecules to move rapidly, which influences various cellular processes.
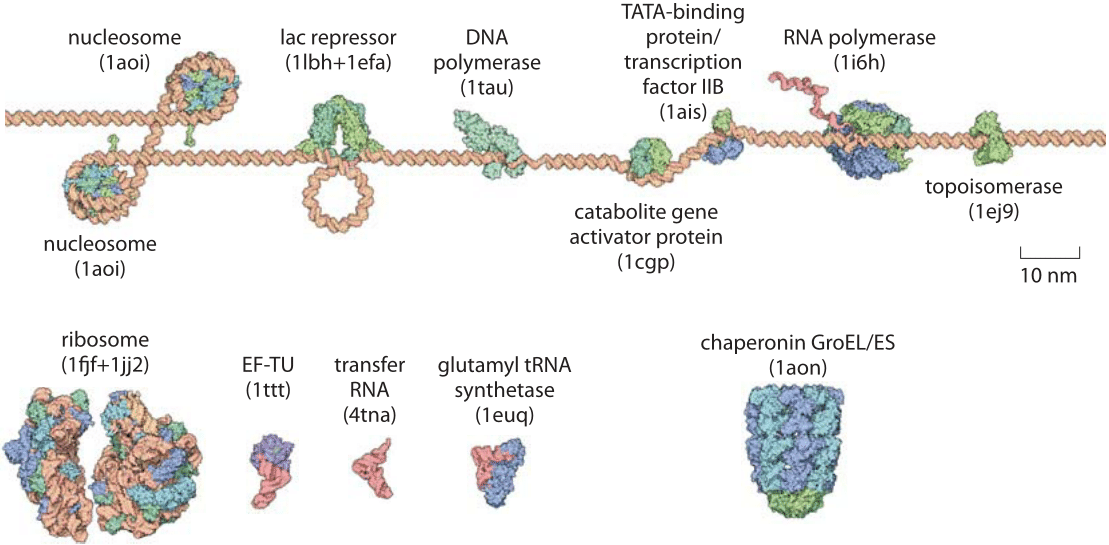 |
|---|
| © bionumbers.com |
Subunits have Distinct Functions (RNAP)
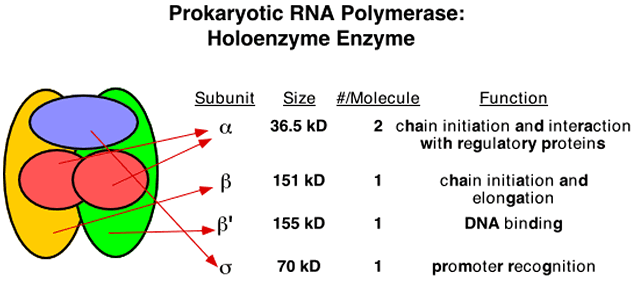 |
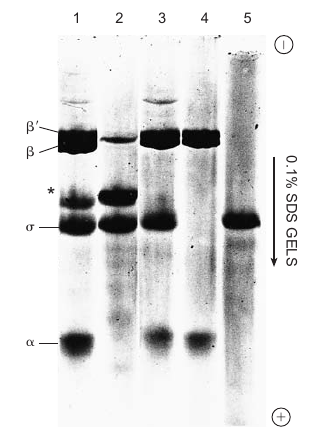 |
|---|---|
| © golifescience | P122; Figure 6.1 |
Initial Steps
- Promoter Search: RNA polymerase locates promoter regions, an ATP-consuming process, to initiate transcription.
- Closed Promoter Complex Formation: RNA polymerase binds to the promoter region without unwinding the DNA double helix.
- Open Promoter Complex Formation: The DNA double helix unwinds, allowing RNA polymerase to initiate RNA synthesis.
$\sigma$ Factors Driving Specificity/Stability
-
Background:
- Core enzyme indiscriminate about the T4 genes it transcribes, it also transcribes both DNA strands.
-
Experiment:
- RNA made by the holoenzyme did not hybridize with authentic T4 RNA, confirming it is made asymmetrically like the in vivo RNA.
- About 30% of the RNA made by the core enzyme hybridized with authentic T4 RNA and became RNase-resistant.
-
Conclusion:
- The core enzyme transcribes both DNA strands, unlike the holoenzyme that transcribes asymmetrically. This suggests the core enzyme’s activity is not as specific as the holoenzyme’s.
-
$\sigma$ factors are essential for promoter recognition and transcription initiation.
-
They stabilize DNA-bound RNA Polymerase, ensuring efficient transcription.
Filter Binding Assay:
-
DNA molecules can pass through a filter, but the $\sigma$ factor cannot. When mixed together, DNA molecules associate with the $\sigma$ factor, preventing them from passing through.
-
$\sigma$ factor aligns RNA Polymerase at a promoter, ensuring accurate transcription initiation.
Assay for Locating the ‘Melted’ DNA:
- Radio labeling is used to tag specific DNA sequences.
- Methylation of DNA occurs at specific sites.
- Cleavage of DNA at methylated sites provides insights into DNA regions that are “melted” or unwound during transcription.
Technics
Filter binding assay
 |
|---|
| P108, Figure 5.35 |
Nitrocellulose Membrane Filters and DNA-Protein Interaction
Background:
Nitrocellulose membrane filters have long been used for filter-sterilizing solutions. They are also known to bind DNA, but under specific conditions.
Key Points:
-
Binding Conditions:
- Single-stranded DNA: Binds readily to nitrocellulose.
- Double-stranded DNA: Does not bind to nitrocellulose by itself.
- Protein: Binds to nitrocellulose. When bound to double-stranded DNA, the protein-DNA complex will bind to the filter.
-
Assay Demonstrations:
- Figure 5.35a: Labeled double-stranded DNA passed through the filter shows that it does not bind to nitrocellulose. All labeled material found in the filtrate.
- Figure 5.35b: Labeled protein is filtered and is found to bind to the nitrocellulose filter. Demonstrates that proteins can bind independently.
- Figure 5.35c: Labeled double-stranded DNA mixed with a binding protein shows that the protein-DNA complex binds to the filter. The radioactivity is found bound to the filter, confirming the interaction.
Conclusion:
Filter binding assays using nitrocellulose membrane filters serve as a direct measure of DNA-protein interaction. This makes nitrocellulose filters a useful tool for studying such interactions.
Methylation-S1 nuclease
 |
 |
|---|---|
| Figure 6.15; P133 | Figure 6.16; P134 |
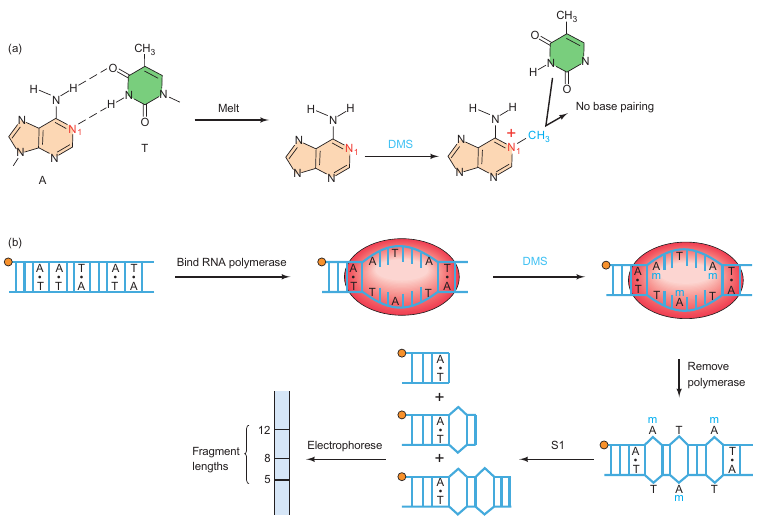
Background:
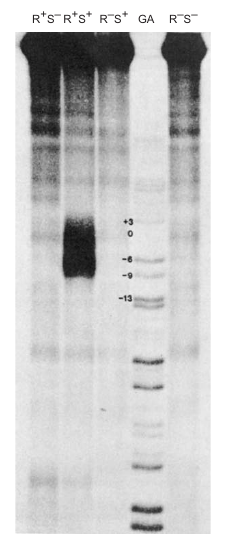
In 1979, Ulrich Siebenlist carried out an experiment to identify the base pairs melted by RNA polymerase in a T7 phage early promoter. The aim was to understand the local DNA melting involved in the formation of an open promoter complex.
Key Points:
-
Experiment Strategy:
- Step 1: End-label the promoter DNA.
- Step 2: Add RNA polymerase to form an open promoter complex, which involves local DNA melting.
- Step 3: Methylate exposed adenines with dimethyl sulfate (DMS).
- Step 4: Remove RNA polymerase to allow the region to close.
- Step 5: Treat the DNA with S1 nuclease, which specifically cuts single-stranded DNA.
- Step 6: Electrophorese the labeled DNA fragments to determine their lengths.
-
Chemical Agent Used: Dimethyl sulfate (DMS) methylates exposed adenines, preventing them from base-pairing with thymines in the opposite strand when the region closes.
-
S1 Nuclease: Specifically cuts where an adenine had been in a melted region and had become methylated.
-
Results:
- A series of fragments extending from position 13 to 29 were observed, rather than a neat set.
- The length of the melted region was estimated to be 12 base pairs.
- G–C pairs were not detected due to the experimental conditions used for methylation.
Conclusion:
The melted region in the T7 phage early promoter was identified to extend from positions 13 to 29 and to have a length of approximately 12 bp. This is in agreement with previous estimates. The experiment revealed that the melted region is precisely where RNA polymerase begins transcribing, which is of biological significance. However, the presence of G–C pairs in the region could not be verified due to limitations in the methylation conditions used.
Advanced Molecular & Cell Biology 1
https://karobben.github.io/2023/08/22/LearnNotes/UIUC-AdMC-1/








