Advanced Molecular & Cell Biology 2
Points for the lecture:
-
Subunits of any complex typically have distinct activities
- The distinct functions of subunits are evident in the RNA polymerase complex, where each subunit specializes in a particular aspect of transcription. For instance, the α subunit is crucial for initiation and interaction with regulatory proteins.
-
Sigma factors, in part, determine promoter specificity
- Sigma factors are essential for guiding the RNA polymerase to specific sets of genes, ensuring the right genes are expressed under certain conditions.
-
Two modes of transcriptional termination in bacteria
- These are the Rho-independent and Rho-dependent mechanisms. Each has its unique sequence cues and processes to halt transcription.
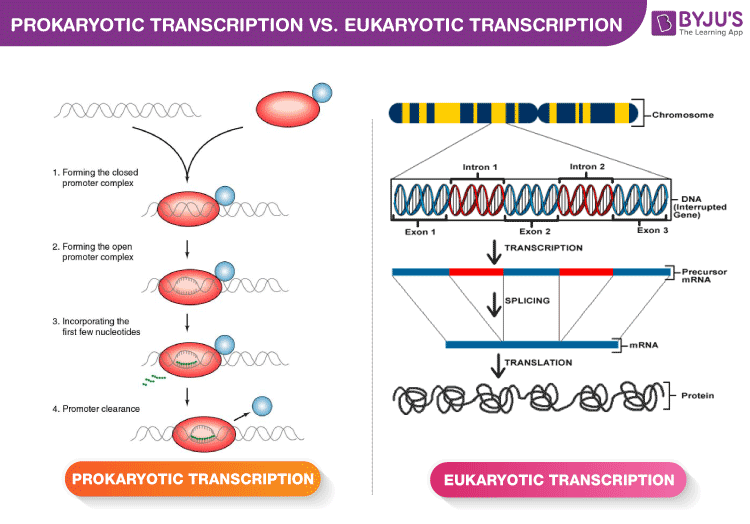
Transcription Initiation In Four Steps
- Forming the closed promoter complex
- This is the initial binding of the RNA polymerase to the promoter region without unwinding the DNA.
- Forming the open promoter complex
- Here, the DNA around the promoter region unwinds, providing a single-stranded template for transcription.
- Incorporating the first few nucleotides
- RNA polymerase begins the synthesis of the RNA strand. It’s a crucial step, ensuring the fidelity of the process.
- Promoter clearance
- After successful initiation, the RNA polymerase moves forward, leaving the promoter region, and enters the elongation phase of transcription.
$\sigma$ Factor is Recycled
After initiation, the sigma factor can dissociate from the RNA polymerase and be used again for another round of transcription initiation. This recycling process is efficient for the cell.
It is discoverd in a very simply assay. After add the fresh core, it got more transcription products.
Red curve: Only one round of chain initiation is possible at low ionic strength because (core) polymerase does not dissociate from DNA.
Green curve: Fresh core polymerase added. (Products surge)
Förster Resonance Energy Transfer (FRET)
experiment: retain the core during the cycle.
“FRET studies have been pivotal in understanding sigma factor dynamics during transcription. By employing FRET, the interaction between sigma factors and the core RNA polymerase during various stages of transcription was elucidated.”
- $\sigma$ Can Remain with Elongating RNAP
- If he doesn’t release, it goes slower.
- By attach the accepter at the end of the gene, once doner from the factor attached the accepter and the light signal changed, which means $\sigma$ factor stay with the DNA.
 |
|---|
| © Eva Stejskalova |
Determing Mechanism(s) of Action aka, Biochemistry
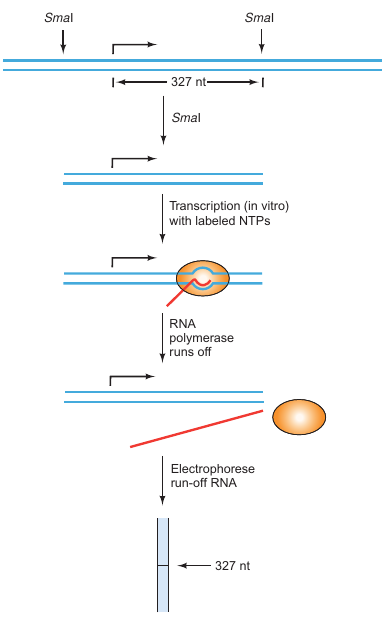
Run-Off Transcription Assay
This assay is designed to assess how quickly RNA polymerase can transcribe a given DNA template. By analyzing the rate and extent of transcription, insights into the efficiency of the transcription process under various conditions can be obtained.
 |
Objective: To mutate a gene’s promoter and observe the effects of mutations on the accuracy and efficiency of transcription. Requirements: |
|---|---|
| P103; Figure 5.31 |
Methodology:
-
Preparation of DNA Fragment:
- Start with a DNA fragment containing the gene you want to transcribe.
-
Restriction Enzyme Cutting:
- Cut the fragment with a restriction enzyme in the middle of the transcribed region.
-
In Vitro Transcription:
- Transcribe the truncated gene fragment in vitro using labeled nucleotides, so the resulting transcript is labeled.
-
Run-Off Mechanism:
- Since the gene has been cut in the middle, the RNA polymerase will reach the end and “run off”, giving the method its name.
-
Measure Transcript Length:
- The length of the run-off transcript confirms the starting point of transcription based on the known location of the restriction site.
Advantages:
- Simplicity: Compared to S1 mapping or primer extension, run-off transcription is simpler.
- Quantitative: It can serve as a quantitative method to measure the rate of in vitro transcription.
Limitations:
- In Vitro Only: The method relies on in vitro transcription and cannot provide information about cellular transcript concentrations.
- Accuracy: It is effective only for genes that are accurately transcribed in vitro.
Applications:
- After identifying the physiological transcription start site through other methods like S1 mapping or primer extension, run-off transcription can be used to quantify transcription rates in vitro.
Summary:
Run-off transcription is a simple, yet effective method for studying the efficiency and accuracy of transcription for a gene, especially in in vitro settings. It is particularly useful for quantitative analysis of transcription rates.
- Test the speed of the transcription
Bound $\sigma$ Factor Promotes RNAP Pausing
- Abortive Initiation Predominates
Background:
Until 1980, it was widely believed that transcription initiation was completed upon the formation of the first phosphodiester bond, connecting the first two nucleotides in the RNA chain.
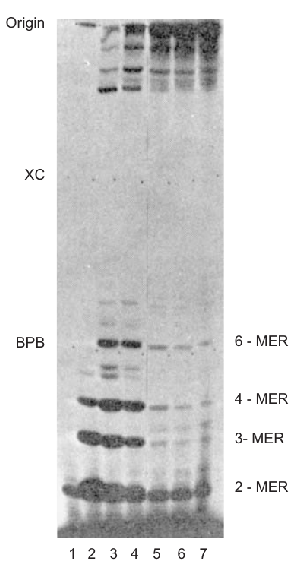
Breakthrough Study by Carpousis and Gralla:
- Experimental Setup:
- E. coli RNA polymerase
- DNA with a mutant E. coli lac promoter (lac UV5 promoter)
- Heparin (prevents reassociation between DNA and RNA polymerase)
- Labeled ATP (to label RNA products)
- Methodology:
- Incubated the polymerase, DNA, heparin, and labeled ATP.
- Ran gel electrophoresis on the products to measure their sizes.
 |
Key Findings: |
|---|---|
| P127; Figure 6.7 |
Implications:
- Abortive Transcripts: The polymerase was producing many small, abortive transcripts without leaving the promoter region.
- Complex Initiation Process: Transcription initiation is more complex than previously thought, involving a stage where multiple abortive transcripts are produced.
- Role of Heparin: Heparin in the assay prevented free RNA polymerase from reassociating with the DNA, confirming that the abortive transcripts were made by the same polymerase without it detaching from the DNA.
Follow-up Research:
Other researchers have corroborated these findings, even discovering abortive transcripts up to 9 or 10 nt in size.
Summary:
The study by Carpousis and Gralla showed that transcription initiation is a complex process involving the generation of multiple abortive transcripts. This groundbreaking work has significantly deepened our understanding of the transcription initiation process in E. coli.
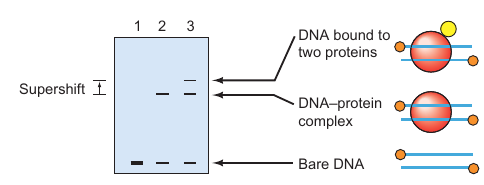
Electro Mobility Shift Assay (EMSA)
 |
|---|
| P109; Figure 5.36 |
Background:
Understanding DNA–protein interactions is crucial for various aspects of molecular biology, including gene regulation. Gel mobility shift assay, also known as electrophoretic mobility shift assay (EMSA), is a technique that allows for the visualization of these interactions.
Principle:
The basic idea is that a small, labeled DNA fragment will move through an electrophoretic gel much more slowly when it is bound to a protein compared to when it is unbound.
Methodology:
- Preparation: A short, double-stranded DNA fragment is labeled.
- Binding: This labeled DNA is then mixed with the protein of interest.
- Electrophoresis: The DNA-protein complex is subjected to gel electrophoresis.
- Detection: The gel is exposed to autoradiography to visualize the labeled species.
Key Findings:
- Naked DNA: Without any bound protein, the labeled DNA shows high mobility in the gel (Lane 1).
- DNA-Protein Complex: When the DNA is bound to a protein, its mobility through the gel decreases significantly (Lane 2). This is the basic “shift” that gives the assay its name.
- Supershift: When the DNA is bound to two proteins, its mobility decreases even further (Lane 3). This is termed a “supershift” and can involve either another DNA-binding protein, a protein that binds to the first protein, or an antibody specific to the first protein.
Applications:
- Study of Protein-DNA Interactions: Allows researchers to understand which proteins can bind to a specific DNA sequence.
- Supershift Analysis: Useful in identifying secondary proteins or antibodies that can further bind to the DNA-protein complex.
- Quantitative Analysis: The degree of shift can also provide quantitative insights into the binding affinities.
Summary:
EMSA is a powerful and versatile technique for studying DNA–protein interactions. It allows not only the qualitative identification of interacting proteins but also offers clues about the binding dynamics through supershifts.
- Radioisotopically Labeled ATP
T4 poly… to tag the DNA end and so, it could create radio-labeled - Only $\beta$’ Subunit of Core RNAP Binds All Promoters
rifampicin actually blocks early elongation
b-subunit from a rifampicin-resistant bacterium, the resulting polymerase was antibiotic-resistant.
b-subunit came from an antibiotic-sensitive bacterium, the reconstituted enzyme was antibiotic-sensitive,
$\beta$ and $\sigma$ Bind to ‘Open’ DNA Strands
- Detecting Protein:DNA Complexes by Crosslinking (Chromatin Immunoprecipitation (ChIP), Figure 5.39)
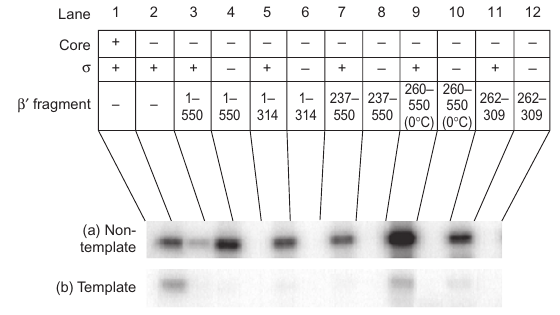 |
The small 262-309 fragment of b9 is critical for stimulating s binding to the non-template DNA strand, and mutations within this fragment significantly interfere with the binding. |
| P142; Figure 6.24 |
- Fragment 1-550: This fragment of b9 induced binding between s and the non-template strand DNA.
- Smaller Fragments: All fragments could induce binding; fragment 260-550 worked only at low temperature.
- Critical Fragment 262-309: A small fragment with 48 amino acids significantly stimulated binding at room temperature.
- Mutations (R275, E295, A302): Known to interfere with s binding to promoters and caused significant interference in binding to the non-template strand in the -10 region.
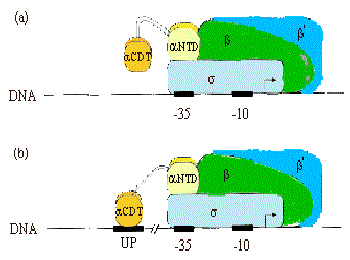
Strong Promoters Have Another Conserved DNA Element (UP element)
 |
 |
|---|---|
| © Dr. Tracy Nixon | P143; Figure 6.26 |
DNase Footprinting
- DNase Footprinting: Uses DNase I to cut unprotected DNA regions.
- Dimethylsulfate (DMS) Footprinting: Uses DMS to methylate adenine bases in DNA.
- Hydroxyl Radical Footprinting: Uses hydroxyl radicals to cleave DNA.
- DNase Footprinting Steps:
- End-Label DNA: Only one strand is labeled per experiment.
- Protein Binding: The target protein is allowed to bind to the DNA.
- DNase I Treatment: Mild conditions used to ensure only one cut per DNA molecule on average.
- Protein Removal: The protein is removed to leave only the DNA.
- Gel Electrophoresis: DNA fragments are separated on a high-resolution gel.
- Control & Multiple Concentrations: A control with DNA alone is included. Different protein concentrations are tested to observe gradual disappearance of bands.
- Outcome:
- The “footprint” region on the gel corresponds to the DNA region protected by the protein, revealing the protein’s binding site on the DNA.
- Significance:
- Helps in identifying the exact DNA regions or bases involved in binding with a specific protein.
-
Strong Promoters Have Another Conserved DNA Element
- -35 box and -10 box. As long as the promoter binding site close to the gene.
-
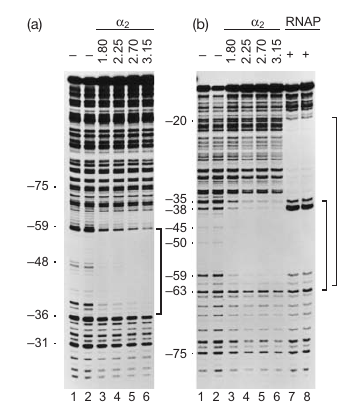
Higher Activity Requires ‘Up’stream Sequence and Carboxyl-terminus of $\alpha$-subunit
- example:
- wilde-type-88 → create lots of DNA; -44: almost makes no DNA
- $\alpha$-235 → never makes lot amount of DNA
Result: $\alpha$ initiation the recognition.
- example:
-
DNA Footprinting
- the position which protein binds the DNA where never be replicated/transcript. As a result, there is a area of the gel lost and know ans the footprint.
-
$\alpha$ Subunit Binds Up Element Independently
-
C-Terminal Domain of $\alpha$ Subunit Can Bind the “Up” Element When Present
Transcription Termination
Termination is an essential step to ensure only the required genes are transcribed and to prepare RNA polymerase for another round of transcription. Both Rho-independent and Rho-dependent mechanisms have unique triggers and processes to ensure efficient termination.
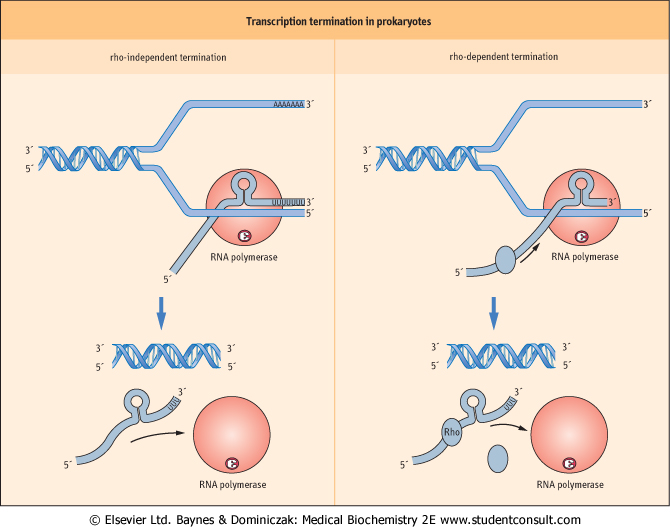
- Only certain regions of a genome are transcribed
- Transcription complexes assemble at promoters and disassemble at the 3’ end of genes at specific termination sequences
- Two types of termination sequences:
- Rho-independent termination (Central dogma forward)
- Formation of an RNA Hairpin: thermodynamic favorite
- Intrinsic Termination Depends on Adenine Run
- Rho-dependent termination (got in the control)
- Rho binds to transcript at rho loading site and pause polymerase
- Hairpin forms; polymerase pauses; roh catches up.
- Rho helices releases transcript and causes termination
- Rho-independent termination (Central dogma forward)
- Initiation or Termination can be Used for Regulation
Significant: In the microbes, a single operon usually activate a serious of protein for a pathway. But we usually only need apart of the genes as a result, the termination study is very important in microbe engineering
Housekeeping vs. Regulated
“Housekeeping genes are essential for the day-to-day functions of the cell and are continuously transcribed. On the other hand, regulated genes are transcribed only under specific conditions. The cell uses various transcription factors and sigma factors to control the expression of these genes.”
- Expression of housekeeping genes is constitutive (essentially)
- Housekeeping genes have strong promoters and are efficiently and nearly continuously transcribed *housekeeping genes whose products are required at low levels have weaker promoters
- Regulated genes are expressed at different levels under different conditions using specific operators/response elements within each promoter
Sigma Factor Directed Transcription Programs
“E. coli, a model bacterium, utilizes different sigma factors to control the transcription of various sets of genes. Depending on the environmental conditions, E. coli can switch between these sigma factors to ensure the appropriate genes are being expressed.”
- Bacterial Gene Regulators
E. coli can choose between 7 sigma factors and about 350 transcription factors to fine tune its transcriptional output
An Rev Micro Vol. 57: 441-466 T. M. Gruber
| Sigma subunit | Type of gene controlled | # of genes controlled |
|---|---|---|
| RpoD | $\sigma^{70}$ Growth/housekeeping | ~1000 |
| RpoN | $\sigma^{54}$ N2; stress response | ~15 |
| RpoS | Stationary phase, virulence | ~100 |
| RpoH | $\sigma^{32}$ Heat shock | ~40 |
| RpoF | Flagella-chemotaxis | ~40 |
| RpoE | ? | ~5 |
| FecI | Ferric citrate transport | ~5 |
[R]poD: means find by gene screening
[P]…: Discovered by Chemical way
Stress: make very unique proteins for survival
Different sigma, different RNA (may other sigmas), Different way (final products)
- Sigma Factors Enable Large Changes in Gene Programs
- Directing Simple and Cascade Responses
Sigma Factor Regulation
“Sigma factors are regulated at multiple levels to ensure appropriate cellular responses. For instance, the sigma factor σ32 is a heat-shock protein. Under normal conditions, it’s unstable and degraded quickly. However, under heat-shock conditions, it’s stabilized, allowing the cell to respond to the stress.”
- $\sigma ^{32}$ is the first $\sigma$ factor bing studied.
- $\sigma ^{32}$ Controls Expression of the Proteins that Control It (a.k.a. Feedback loop)
- $\sigma ^{32}$ Is Continuously Produced but Rapidly Degraded
- $\sigma ^{32}$ Regulation is a Balanced Sensor
- HSPs Reactivate $\sigma ^{70}$ During Recovery
!!! ? what happened between $\sigma ^{32}$ and $\sigma ^{70}$? How cell make the choice?
- $\sigma ^{32}$ is a heat-shock protein. It would degreedated without heat-shock. Heat-shock genes stabilized the $\sigma ^{32}$. In this case, the $\sigma ^{32}$ could win the competition which $\sigma ^{70}$ DnaK, DnaJ, and GrpE are heat-shock proteins work to protect the $\sigma ^{32}$.
Advanced Molecular & Cell Biology 2
https://karobben.github.io/2023/08/23/LearnNotes/UIUC-AdMC-2/