Molecule and Cellular Biology 3
Reading for this week:
- Book
Molecular Biology Chapters 7.1, 7.2 - Review:
Transcription activation by catabolite activator protein (CAP). Busby, S. and Ebright, R.H. J Mol Biol. 1999 Oct 22;293(2):199-213.
Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Dillon, S.C. and Dorman C.J. Nat Rev Microbiol. 2010 Mar;8(3):185-95. - Journal Club Paper (to be presented the following week):
A phage-encoded nucleoid associated protein compacts both host and phage DNA and derepresses H-NS silencing. Son B, Patterson-West J, Arroyo-Mendoza M, Ramachandran R, Iben JR, Zhu J, Rao V, Dimitriadis EK, Hinton DM. Nucleic Acids Res. 2021 doi: 10.1093/nar/gkab678.
Points for the lecture:
- Transcription can be actively regulated
- Small molecules can control gene programs
- Operons help balance protein expression in a pathway
- co-regulate as unite
- Protein allostery
- Many level, 1. small molecular, 2. DNA (Bind something to make conformation change.)
- Transcription factors can bend DNA
Role of Regulatory Proteins in Transcription Initiation
- Regulatory proteins bind to specific DNA sequences (operator/response element) and control initiation of transcription
- Repressors - regulatory proteins that prevent transcription of a negatively regulated
gene *negatively regulated genes can only be transcribed in the absence of the repressor - Activators - regulatory proteins that activate transcription of a positively regulated gene
- Repressors and Activators are often allosteric proteins modified by ligand binding
- Agonist - positively acting ligand
- Antagonist - negatively acting ligand
Activator Strategies for Transcriptional Regulation
-
Ligand binding triggers promoter association and RNA is produced (Trigger the initiation)
-
Ligand binding triggers promoter dissociation and RNA is not produced (Stop the transcription)
THe contact is so much and some of the mechanism are still not figured out.
Repressor Strategies for Transcriptional Regulation
- Ligand binding triggers promoter dissociation and RNA is produced
- Ligand binding triggers promoter association and RNA is not produced
Transcription Factors Bind Operators (bacteria)/Response Elements (eukaryotes)
Eu: Inhancers, depends on different regulation gene, far away from the genes.
Ba: almost all close to the gene
!!! How do you find an Operator/Response Element?
- Operators
- Many TFs bind inverted repeats as dimers
- More interactions = greater affinity
- Enhancer
- Promoter ‘Bashing’
lacZ could be a report aod transcription reporter and change the color of the cell in this assay.
- Linker-scanner Mutagenesis to Refine Position of Promoter Proximal Elements (after the Promoter ‘Bashing’)
peron
Bacteria and viruses often express functionally operative genes from a single promoter to help insure
uniform expression
DNA order: Activator binding site promoter, Repressor binding sit (Operator)
Example: Lactose (Lac) Operon
- Lactose permease (Lacy)
- B-Galactosidase (LacZ)
- Thiogalactoside transacetylase (LacA)
!!! What about the Lactose operon:
The lac operon in E. coli is one of the most well-studied systems for gene regulation. This operon contains genes for enzymes required for the metabolism of lactose, a disaccharide sugar. The operon consists of three genes:
- **lacZ**: Codes for \(\beta\)-galactosidase, which hydrolyzes lactose into glucose and galactose.
- **lacY**: Codes for galactoside permease, a transport protein that facilitates the entry of lactose into the cell.
- **lacA**: Codes for galactoside transacetylase, whose function is not entirely clear.
These genes are transcribed into a single polycistronic mRNA, meaning that they are expressed together. This allows for coordinated control of these functionally related genes.
- **Negative Control: The Brake**
In the absence of lactose, a protein called the lac repressor binds to the operator region of the operon, blocking RNA polymerase and thus preventing transcription. This is a form of negative control; it keeps the operon turned off when lactose is not present, which is efficient for the cell.
- **Positive Control: The Accelerator**
The lac operon also has a system for positive control, mediated by an activator protein that responds to glucose levels in the cell. This activator is cAMP-CAP (cAMP Receptor Protein). When glucose levels are low, cAMP levels are high, allowing CAP to bind and enhance the transcription of the lac operon.
- **Diauxic Growth: Switching from Glucose to Lactose**
When E. coli is grown in a medium containing both glucose and lactose, it exhibits diauxic growth. The bacteria consume glucose first and only switch to lactose metabolism once glucose is depleted. This switch involves a lag phase where the lac operon is activated, allowing the cells to make the enzymes needed for lactose metabolism.
- Advantages of Dual Control
1. **Economic Efficiency**: Negative control ensures that the enzymes for lactose metabolism are not produced when lactose is not present. This is energy-efficient.
2. **Resource Prioritization**: Positive control ensures that even if lactose is present, the lac operon will not be fully activated if glucose is also available. This allows E. coli to prioritize the easier-to-metabolize glucose.
In summary, the lac operon is a sophisticated system that allows E. coli to adapt to changing nutritional conditions efficiently, ensuring that it utilizes available resources in a manner that is both effective and economical.
The Operon Model (History)
- A landmark in the history of Molecular Biology was the proposal of the operon model of genetic regulation by François Jacob and Jacques Monod in 1961.
- The core of the model was that the level of proteins in cells was controlled at a genetic level. The theory also predicted the existence of mRNA-an unstable intermediate between the genome and the expressed protein.
- Before this work the prevailing model was called the instruction hypothesis that stated that all proteins were present in a cell, but that in the absence of an inducer they were not properly folded and were inactive.
- Jacob and Monod were awarded the Nobel Prize in 1965 for their work on characterising genes involved in lactose utilization
Differential Growth based on Carbon Source
-
Glucose is more rapidly used than lactose, so cells grow faster on glucose.
-
When both present growth is bi- phasic, which is called a diauxic shift.
When you mix up them, the bacteria make the choice. They know when and how to switch the strategies.
Cell numberBiological Purpose of Lac Operon
-
LacZ gene: encodes $\beta$ -galactosidase enzyme that breaks down disaccharide lactose into glucose and galactose to be used as source of energy for cell.
-
LacY gene: encodes $\beta$ -galactoside permease, a membrane-bound protein that helps transport lactose across the membrane of cell.
-
LacA gene: encodes -galactoside transacetylase enzyme involved in catabolism of disaccharide (exact role is less clear)
- Genetic Screen Based on Enzymatic Activity (Blue: lacZ activity)
5-bromo-4-chloro-3-indolyl-$\beta$D-galactoside- Natural & Synthetic Small Molecules
IPTG is a non-metabolized analog of allolactose that induces Lac operon but is not broken down by enzymes.
Lactose is a disaccharide composed of galactose and glucose linked in a configuration (1->4). -galactosidase, the product of the lacZ gene, cleaves this disaccharide into galactose and glucose which can then both be metabolized by the cell. E. coli does not always make this enzyme. It is an inducible enzyme which is synthesized only when lactose is present and when glucose is absent from the growth media. Glucose is one of the few sugars which E. coli will use preferentially over all other sugars because it can be directly routed into glycolysis after phosphorylation without any other modifications.
Merodiploids Helped to Unravel the Role of Genes in the lac operon
Studying the Lac Operon
Isolation and analysis of mutants defective in Lac operon function allowed Jacob and Monod to formulate an accurate Lac operon model PRIOR to the advent of molecular cloning
e.g., wild type E. coli form colonies that are blue on IPTG/ XGAL plates and white on XGAL plates
Mutants that form white colonies on IPTG/XGAL plates can be isolated * this phenotype is often due to a mutation in a Lac structural gene * the number and location of Lac structural genes can be identified by complementation analysis
Mutants that form blue colonies on XGAL (no IPTG) can be isolated * this phenotype is often due to a mutation in either the Lac repressor gene or Lac operator * Lac repressor versus operator mutations can be distinguished by cis-trans test F’ plasmids carrying different pieces of E. coli chromosome can be isolated in HFR strains, allowing the creation of partial diploids.
F’ plasmid can carry wild type or mutant versions of Lac operon between strains
Complementation Analysis and Cis-Trans Tests
Phenotypes and Genotypes of Mutants Defective in Lac Operon Function
- Lower case letter with superscript asterisk indicates location of mutation
- In practice, you would not know the location; you would only know the phenotypes (blue or white color on XGAL/IPTG or XGAL) of the haploid (or homozygous diploid) and the heterozygous diploid
e.g., The diploid resulting from conjugating mutants 2 and 3 give a normal phenotype, consistent with mutations 1 and 2 being recessive and located in different structural genes (i.e., in the diploid there is at least one normal copy of each gene and a correspondingly normal protein)
Mutaion -> got the different phynotype -> trans is protine, cis is DNA. -> Bind the two mutation: Make the crosses and repeat the phenotype. They don’t know what are they yet. The name is made by them and the mechanim are solved years later after the sequencing techs.
‘Trans’ and ‘Cis’
-
In genetic terminology: The lac repressor is a trans-acting element
trans = across -
The operator is a cis-acting element
cis=near
The lac repressor is a sequence-specific DNA-binding protein that Negatively regulates expression of the lac operon structural genes.
Trans Complementation
Merodiploid: lacZ - / F’ lacY -
lacI
lacO lacZ
lacY
lacA
lacI
lacO
lacZ
lacY lacA
‘Simple’ Mechanism for Gene Regulation by Lac Repressor
 |
|---|
| P170; Figure 7.3 |
- Negative Control
- Operon naturally “on.”
- Turned “off” by repressor protein from lacI gene.
- Allosteric Regulation
- Repressor is allosteric; its shape changes.
- Inducer is allolactose, changes repressor shape.
- Changed repressor can’t bind to operator, operon “on.”
- Basal Level & Initial Activation
- Repression “leaky,” small amounts of β-galactosidase and permease always present.
- Enough for initial lactose uptake and allolactose creation.
- Positive Feedback (Snowball Effect)
- Derepression leads to more β-galactosidase and permease.
- More lactose uptake and allolactose creation.
- Operon stays “on” if lactose present and glucose absent.
- Summary
- Negative control via repressor.
- Allosteric regulation via allolactose.
- Initial activation via “leaky” repression.
- Positive feedback maintains activation.
Lactose Levels “Tune” the Response
In the absence of lactose there are only 1-5 molecules of each of the lacZYA proteins in the cell. Following
induction, up to 5000 molecules of -
galactosidase can accumulate within
minutes.
As lactose is cleaved and used as a carbon source for growth the lactose levels drop. Eventually free repressor molecules will accumulate once more and the lac operon will be switched off.
Impact of Inducer on Repressor DNA binding
 |
|---|
| P174; Figure 7.6 |
- Challenges Faced
- The lac repressor is in very low concentration in the cell.
- No easy assay to identify the protein was available.
- Strategies Used
- Used a mutant E. coli strain with repressor mutation (lacI t).
- This mutation made the repressor bind to IPTG (isopropylthiogalactoside) more tightly.
- This allowed for detection of the repressor in very impure extracts.
- Purification
- Gilbert and Müller-Hill were able to purify the protein because they could now detect it.
- Operator-Binding Studies
- Conducted by Melvin Cohn and colleagues.
- Used nitrocellulose filter-binding assay.
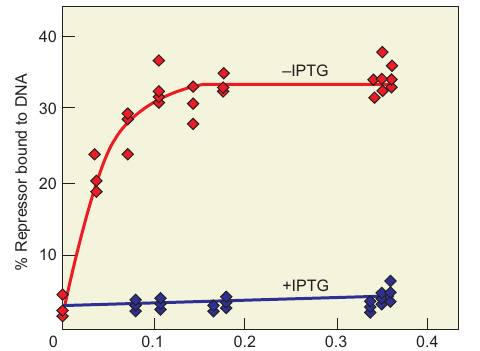
- Results
- Typical saturation curve observed for repressor–operator binding in the absence of inducer.
- No binding observed in the presence of the synthetic inducer, IPTG.
- Summary
- Gilbert and Müller-Hill successfully purified the lac repressor using a mutant strain.
- Cohn’s team demonstrated that inducer (IPTG) blocks repressor–operator binding.
Protein Allostery (Induced Conformational Changes)
Lac Repressor Does Not Prevent RNAP Binding
 |
 |
|---|---|
| Unknow sours | P175; Figure 7.8 |
-
Traditional Assumption: Assumed that lac repressor denies RNA polymerase access to the promoter.
-
Contradictory Evidence
- Pastan’s 1971 Experiment: Showed that RNA polymerase could bind to the lac promoter even in the presence of repressor.
- Straney and Crothers (1987): Showed that polymerase and repressor can bind together to the lac promoter.
-
Alternative Explanations
- Straney and Crothers: Suggested repressor blocks the formation of an open promoter complex.
- Krummel and Chamberlin: Proposed that repressor blocks the transition from the initial transcribing complex to the elongation state, trapping the polymerase in a nonproductive state.
-
Supporting Evidence
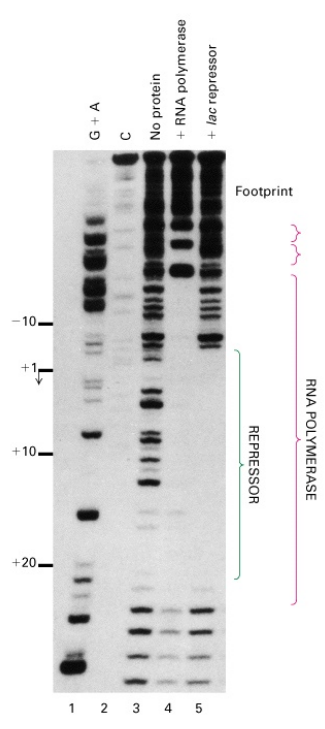
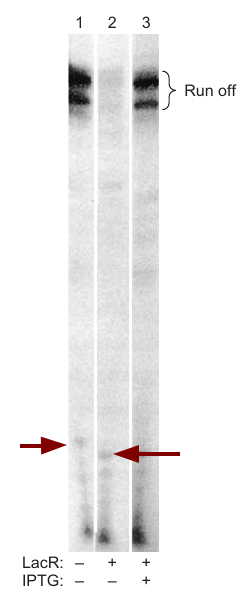
- Jookyung Lee and Alex Goldfarb’s Experiment
- Used a run-off transcription assay.
- Found that RNA polymerase is engaged on the DNA template even in the presence of repressor.
- Noted the appearance of shortened abortive transcripts (about 6 nt long) in the presence of repressor, suggesting repressor may lock the polymerase into a nonproductive state.
- Jookyung Lee and Alex Goldfarb’s Experiment
-
Summary
- Repressor doesn’t seem to block RNA polymerase from accessing the lac promoter.
- Evidence suggests that the repressor may lock the polymerase into a nonproductive state, limiting it to making only abortive transcripts.
What is the Mechanism of Repression?
Influence of Regulators on RNAP:Promoter Complexes
$$
R + P \underset{K_ B}{\rightleftharpoons} RP_ c \underset{k_ 2}{\rightarrow} RP_ o
$$
- R is RNA polymerase
- P is the promoter
- RPc is the closed promoter complex
- RPo is the open promoter complex.
- KB
- Definition: Indicates RNA polymerase’s binding affinity to a promoter.
- Influenced by: Repressors or activators affecting RNA polymerase binding.
- k2
- Definition: Represents the rate-limiting step in forming an open complex; its physical meaning can vary between promoters.
- Influenced by: Repressors or activators affecting RNA polymerase-promoter interactions.
- Insights
Understanding KB and k2 helps in deciphering the effects of auxiliary proteins on RNA polymerase-promoter dynamics. It can hint at how the binding affinity is influenced and the complexities involved in open complex formation.
Impact of Repressor on RNAP
At the lacUV5 promoter, the values of KB and k2 in the presence or absence of repressor are:
| Condition | KB (M⁻¹) | k2 (s⁻¹) |
|---|---|---|
| No repressor | 1.9 x 10⁷ M⁻¹ | 6.7 x 10⁻² s⁻¹ |
| With repressor | 2.5 x 10⁹ M⁻¹ | 2.5 x 10⁻² s⁻¹ |
- Data from Straney & Crothers (1987) Cell 51: 699-707.
- The UV5 mutation changes the -10 region of the lacP promoter so that it more closely resembles the consensus -10 sequence. It is a stronger promoter than lacP.
These data indicate that the lactose repressor INCREASES the binding of RNAP 130-fold; suggest that repressor might be considered a activator. Repressor DECREASES k2 3-fold.
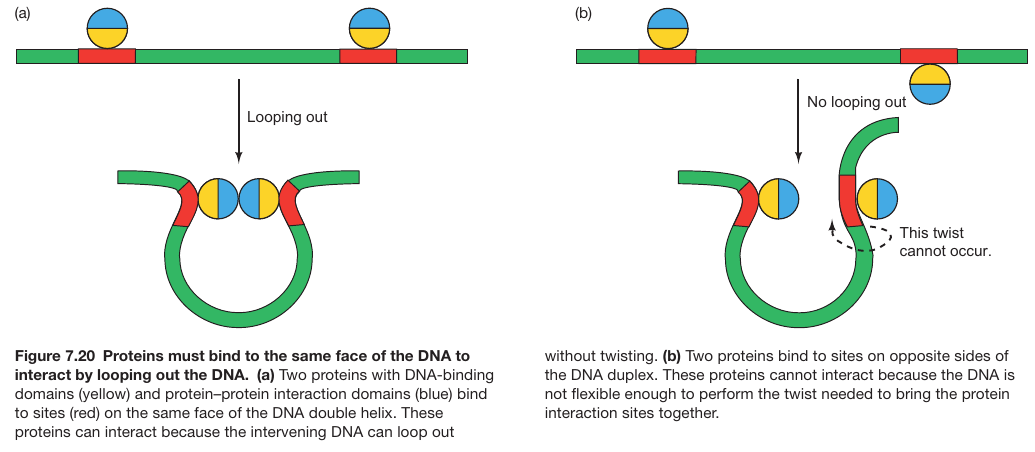
Repressor Dimers Bind on Same Face of DNA
 |
|---|
| Unknown |
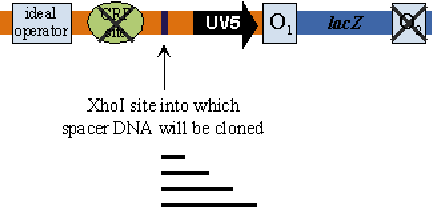
By insert the DNA fragment between the operator and UV5, the transcription efficiency is changed.
- Key Findings:
- Repression maxima: Occur at distances integral to the DNA double helix pitch (59.5, 70.5, 81.5, 92.5, 115.5, 150.5 nm).
- Wild-type lac operon: Has a 92.5 nm spacer distance between operators.
- Gal operon: Features a 115.5 nm spacer distance between operators.
- Longer distances: Display smaller effects, with distances over 400 bp having little to no impact.
- Non-integral multiples: No repression observed, aligning with the DNA looping model’s predictions.
- Implications:
- DNA Looping Model: The observations support a model where interaction is easy when proteins are bound on the same DNA face without needing torsional changes.
- DNA Pitch: Estimated to be 11.1 bp or 3.4 nm in vivo, based on repression maxima data.
- Limitations: Distances under 59.5 nm could not be measured due to interference with the promoter.
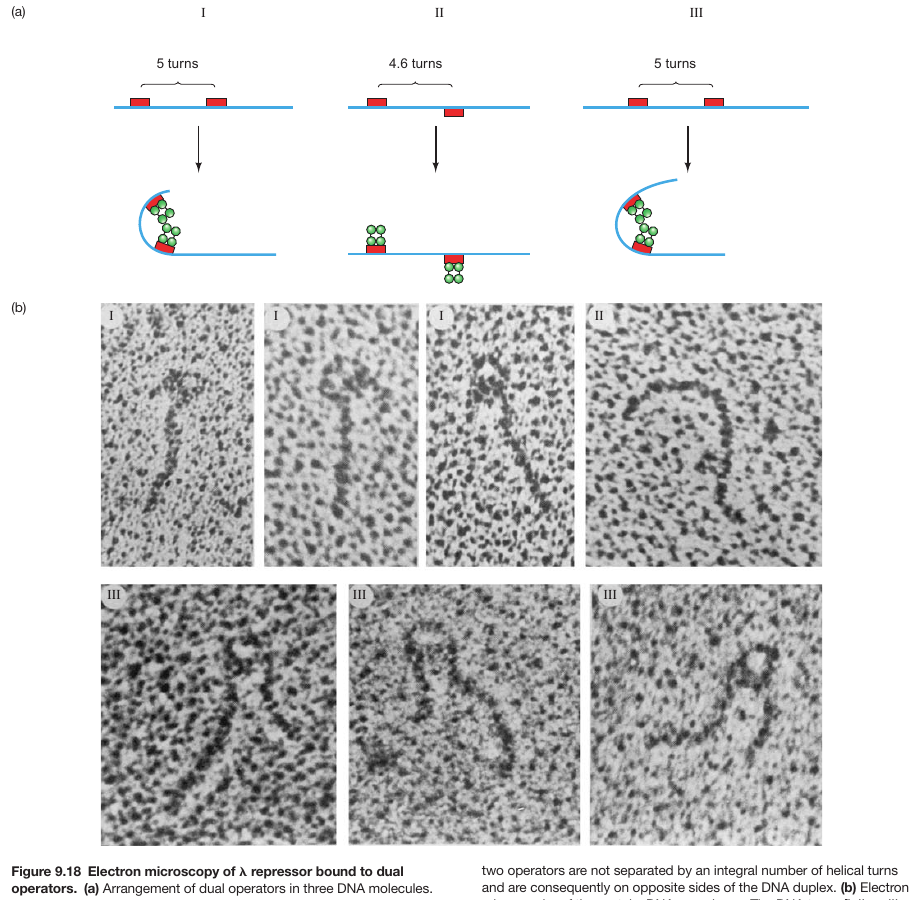
Repressor Binds as a Tetramer
DNA Bending Enables Proteins to Bind Cooperatively to Separated Sites
 |
|---|
 |
| Molecular Biology; Weaver, Robert: Fifth edition |
bending
- BEND FORMATION REQUIRES THAT BOTH PROTEINS BIND TO THE SAME FACE OF THE DNA
No bending
- PROTEINS BOUND TO SITES ON OPPOSITE SIDES OF DNA HELIX CANNOT INTERACT
Lac Tetramer Stabilizes Bent DNA
Detected by EMSA
Gel electrophosphate
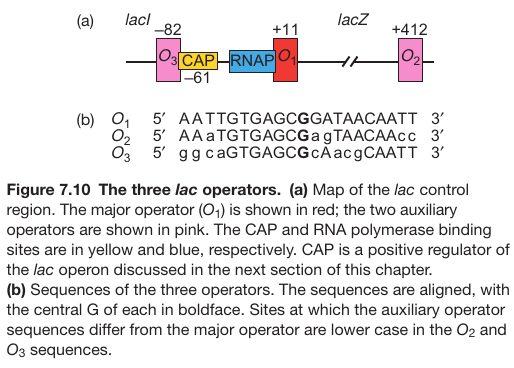
Promoter Contains 3 Operator Sites
 |
 |
|---|---|
| P176; Figure 7.10 | P176; Figure 7.11 |
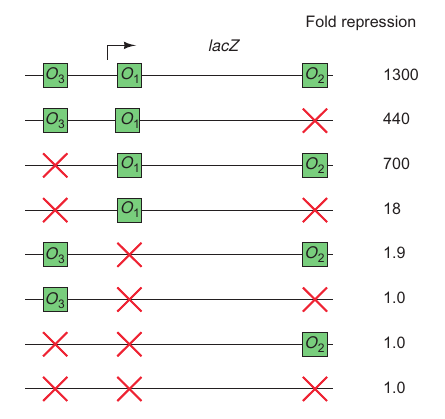
How to Resolve if Operator 2 or 3 is preferred?
- Chromatin Immunoprecipitation (ChIP) Assay
- Determines the relative DNA Occupancy of a Target
- Protein at a Particular Site in vivo
ChIP-Seq can Determine Genome-wide Binding Site Useage of a Target Protein
Operator 1 + Operator 2: bind different place of the DNA and bind the DNA. Then Promoter is loaded much easier.
Lac Repressor Binding
to be next: Multiple Regulators
Molecule and Cellular Biology 3
https://karobben.github.io/2023/08/29/LearnNotes/UIUC-AdMC-3/








