Molecule and Cellular Biology 4
cAMP Levels Control ‘Catabolite Repression’
Catabolite Activator Protein: Ira Pastan and his colleagues demonstrated that cAMP, added to bacteria, could overcome catabolite repression of the lac operon and a number of other operons, including the gal and ara operons.
A protein involved in gene regulation was discovered and named CAP by Zubay and CRP by Pastan’s group. We will use the name CAP to refer to this protein and crp as the gene.
CAP mechanism:
-
Mutation Discovery: Zubay’s team found mutations in the lac promoter preventing CAP and cAMP from boosting lac transcription.
-
Binding Site Location: Research pinpointed the CAP-cAMP binding site upstream of the lac promoter.
-
CAP-cAMP Complex Formation: cAMP modifies CAP’s shape, increasing its affinity for the binding site.
-
Open Promoter Complex Assistance: The CAP-cAMP complex helps RNA polymerase to form an open promoter complex, aiding transcription initiation.
-
Role in Transcription: The complex aids in efficient lac operon transcription, helping the cell adapt to nutritional changes.
In this process, the CAP-cAMP complex facilitates the initiation of transcription in response to environmental cues, enhancing the cell’s adaptability.
- Regulation: many layers
- Positive Reg: alpha-subunit of RNAP
- DNA bending
- Weak promoters
CAP: Catabolic activator protein
repressor responsible to low level of Lactose so it wound transcript when Lactose is low
CAP responsible to the low level of Glu and help the transcription
When Glu is low and lactose is high, the repressor is cleared and CAP is on. The transcription is started.
CAP is a receptor to bind the cAMP. cAMP-CAP complex contribute the activation of the gene expression.
CAP Binding Induces DNA Bending
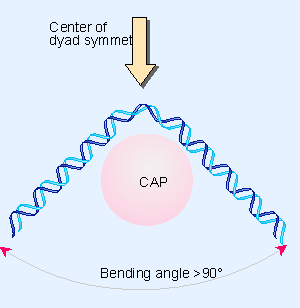 |
|---|
| Figure 10.27 CAP bends DNA >90° around the center of symmetry. |
-
Study Focus: Investigation of the DNA bending effect due to CAP–cAMP binding using electrophoresis.
-
Method:
- Prepared equal length DNA fragments of the lac operon with varied CAP-binding site positions.
- Bound CAP–cAMP to each fragment.
- Performed electrophoresis to observe the migration rates of the DNA-protein complexes.
-
Findings:
- DNA fragments exhibited different migration rates, indicating DNA bending.
- The degree of bending was estimated to be around 90 degrees, aligning reasonably with the 100 degrees found in later x-ray crystallography studies.
-
Implication: The observed DNA bending is believed to facilitate optimal interactions between proteins and DNA in the complex.
CAP-cAMP Binding Functionally Similar to Up Element
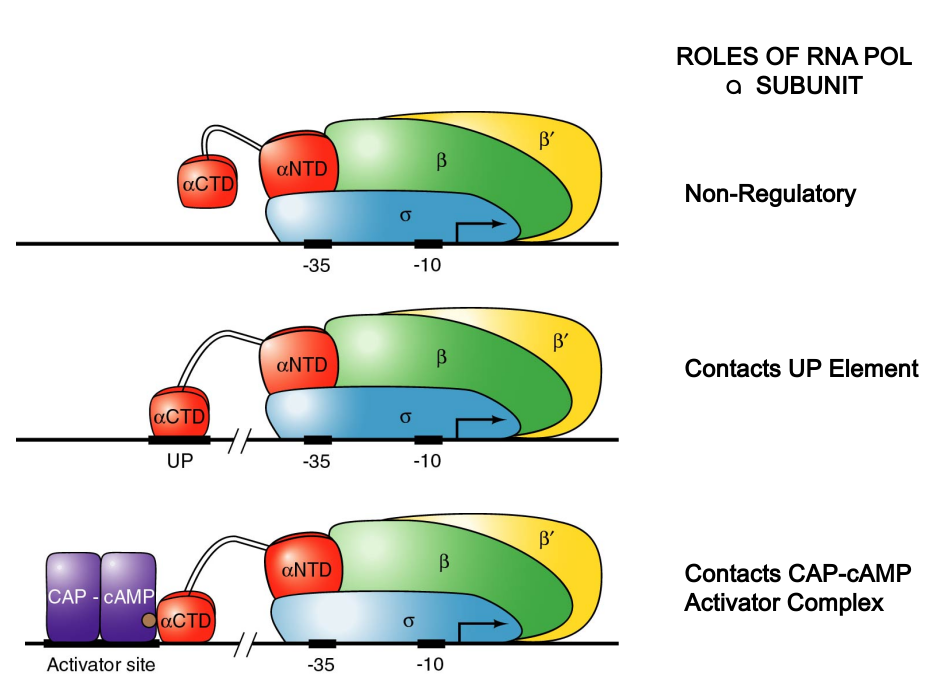 |
|---|
| P182; Figure 7.19 |
-
Study Focus: Importance of interaction between CAP and RNA polymerase’s aCTD in transcription stimulation.
-
Experiment Details: Used RNA polymerases with both normal and truncated a-subunits to transcribe cloned lac operons in vitro.
-
Findings:
- Truncated a-subunits could function like wild-type in absence of CAP-cAMP.
- Truncated a-subunits weren’t stimulated by CAP-cAMP, indicating the necessity of aCTD for CAP-cAMP stimulation.
-
Hypothesis: CAP-cAMP dimer, binding to its activator site, interacts with aCTD, enhancing polymerase’s affinity for the promoter, similar to aCTD’s role with UP elements.
CAP promoters
3 Classes of CAP Regulated Promoters
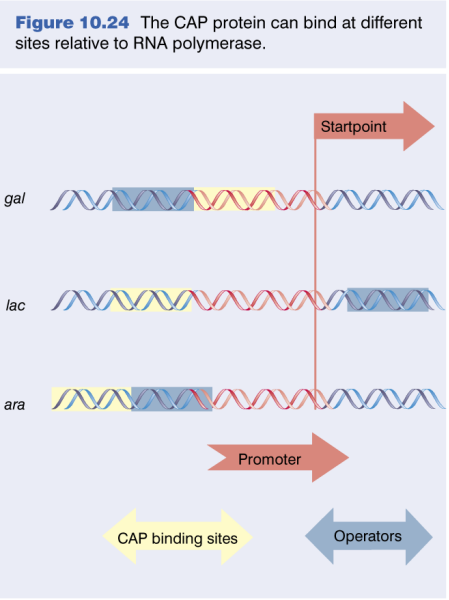 |
|---|
| Unknown |
| Attribute | Class I | Class II | Class III |
|---|---|---|---|
| Activation | Requires only CAP | Requires only CAP | Requires CAP and additional regulators |
| CAP/CRP Binding Site | Upstream of the promoter | Overlaps the promoter, replacing -35 region | Variable, typically >90 bp upstream |
| Prototype | LacP1 | GalP1 | None |
| Location (from start point) | Centered 61.5 bp upstream | Centered 41.5 bp upstream | More than 90 bp upstream |
| Examples | - | - | AraBAD, malK promoter |
CAP Operators Display Periodicity: The greatest activation was seen at distances of -41.5 and -61.5 which correspond exactly to the spacings observed in CLASS II and CLASS I CRP-activated promoters, respectively.
DNA Bending as a Regulatory Mechanism
| Illustrate | Description |
|---|---|
| - Promoter bashing of a hypothetical two-region promoter. The promoter is cloned upstream of the lacZ reporter gene. Point mutations that inactivate each region are made (the red Xs) and the region is cloned onto a plasmid and inserted into E. coli cells, grown up, and has the presence of reporter measured. The binding of Protein B in this example is necessary for RNA polymerase to bind and initiate transcription. - In a laboratory setting, it may not be known that the promoter consists of two regions – single mutations can be made along the promoter, the promoter can be sequenced, and the levels of reporter assayed to find boundaries for each region. |
|
| © wiki |
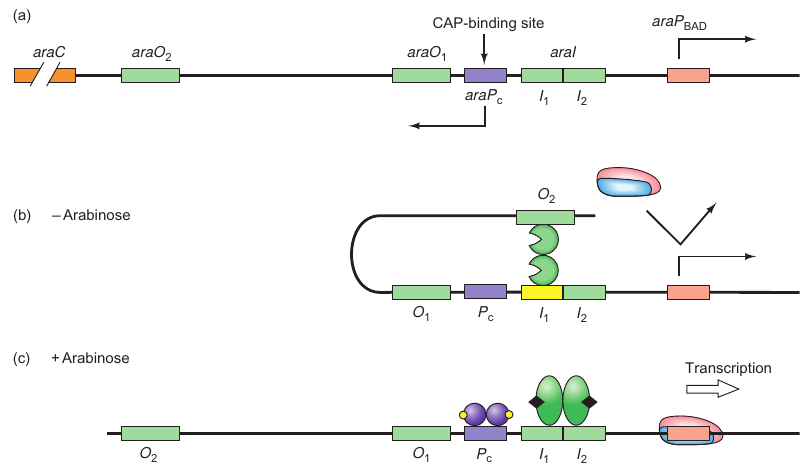
araBAD Operon
- araBAD encodes enzymes responsible for arabinose breakdown
- Operon is catabolite repressed, and thus requires CAP.cAMP for activation of transcription
- Activation also requires the complex of AraC. arabinose as an activator
- Explains why araBAD is only transcribed in the presence of arabinose
Components:
- araO2: Far upstream binding site for AraC.
- araO1: Located between positions -106 and -144, another binding site for AraC.
- araI: Divided into araI1 (-56 to -78) and araI2 (-35 to -51), each can bind one monomer of AraC.
- araA-D: Four genes in the ara operon where araB, araA, and araD are for arabinose metabolizing enzymes and araC is for encoding the control protein AraC.
- araPBAD: Promoter for rightward transcription of araB, A, and D.
- araPC: Promoter for leftward transcription of araC.
States:
-
Without Arabinose (Repressed State):
- AraC binds to araO2 and araI1.
- Causes DNA looping between the binding sites, repressing the operon.
-
With Arabinose (Derepressed State):
- Arabinose alters AraC conformation.
- AraC binds to araI1 and araI2, breaking the repression loop.
- The operon is derepressed.
Positive Control:
- Involves CAP and cAMP.
- They bind to a site upstream of the araBAD promoter.
- DNA looping facilitates CAP’s contact with polymerase, enhancing its binding to the promoter, thereby stimulating transcription.
Figures:
- Figure 7.21a: Illustrates the three binding sites of AraC.
- Figure 7.21b: Shows the repression loop in the absence of arabinose.
- Figure 7.21c: Depicts the derepressed state with the presence of arabinose and how CAP-cAMP complex influences transcription.
This structure helps in detailing the different components involved and the states of the ara operon based on the presence or absence of arabinose, along with the role of CAP and cAMP in its regulation. It also mentions the figures that visualize these processes.
Arabinose Metabolism System
Promoter Bashing Identified Two Operator Sites for Repression
DNA Bending Requires both araO2 and araI Sites
How can araO2 control transcription from a promoter over 250 bp downstream?
The most reasonable explanation is that the DNA in between these remote sites (the operator and the promoter) loops out as illustrated in
Sure! Here is a summarized version of the passage in note form:
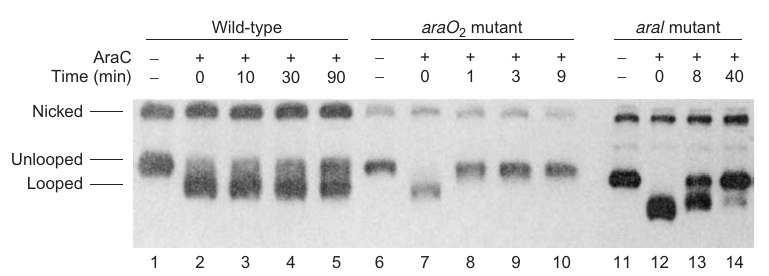 |
|---|
| P184; Figure 7.22 |
Experiment setup:
- Objective: Verify AraC-induced DNA loop formation in the ara operon in the absence of arabinose.
- Materials:
- 404-bp supercoiled minicircle DNA containing araO2 and araI sites, 160 bp apart.
- AraC protein.
- Method: Electrophoresis to measure DNA loop formation by monitoring changes in electrophoretic mobility.
Findings:
-
AraC induces loop formation:
- Evidence: A new high-mobility band observed upon addition of AraC, indicating looped minicircle (Figure 7.22, compare lanes 1 and 2).
-
Loop stability depends on both araO2 and araI:
- Tested using:
- Wild-type minicircle.
- Minicircle with mutant araO2.
- Minicircle with mutations in both araI sites.
- Results:
- Wild-type: Half-time of dissociation ~100 min (50% conversion to unlooped form in 90 min, lanes 3-5).
- araO2 mutant: Loop broke in <1 min, indicating a crucial role in loop maintenance (lanes 7 and 8).
- araI mutant: Half-time of loop breakage <10 min, underscoring its role in loop stability.
- Tested using:
Conclusion:
- AraC can induce DNA looping at the ara operon in the absence of arabinose.
- Both araO2 and araI sites are vital for loop stability; mutations in either site destabilize the loop significantly, showcasing their roles in the AraC-mediated DNA looping mechanism.
Spacing is Important
AraC Binding to araO2 is Disrupted by Arabinose Binding & Regulation of araPBAD
 |
|---|
| P184; Figure 7.21 |
Experiment Setup:
- Objective: Understand the effect of arabinose on the AraC-induced DNA loop and elucidate the role of araI2 in AraC binding.
- Materials:
- Looped minicircles (404-bp supercoiled DNA with araO2 and araI sites).
- Arabinose.
- AraC protein.
- Methods:
- Electrophoresis to observe the influence of arabinose on loop stability.
- Methylation interference to study AraC contacts with araI sites in different states.
Findings:
-
Arabinose breaks the repression loop:
- Evidence: Disappearance of the looped DNA band upon adding arabinose before electrophoresis (refer to Figure 7.23).
-
Reformation of broken loop:
- Conditions: Removal of arabinose allowed the broken loop to re-form, illustrating reversible control mediated by arabinose.
-
AraC’s contact shifts from araO2 to araI2:
- Evidence:
- Methylation interference: AraC contacts araI1 but not araI2 in the looped state. Two araI1 bases were lightly methylated in looped DNA and heavily methylated in unlooped DNA, indicating crucial roles in looping.
- Mutation study: AraI2 mutations didn’t affect AraC binding in looped state but significantly influenced it in the unlooped state, highlighting its role in AraC binding post-loop breakage.
- Evidence:
Conclusion:
- Arabinose disrupts the AraC-induced DNA loop by altering AraC’s affinity for binding sites: it loses affinity for araO2 and gains affinity for araI2, a dynamic regulatory mechanism controlling the ara operon (depicted in Figure 7.21b and c).
- The role of araI2 is crucial in AraC binding in the unlooped state, facilitating the reformation of the loop upon arabinose removal.
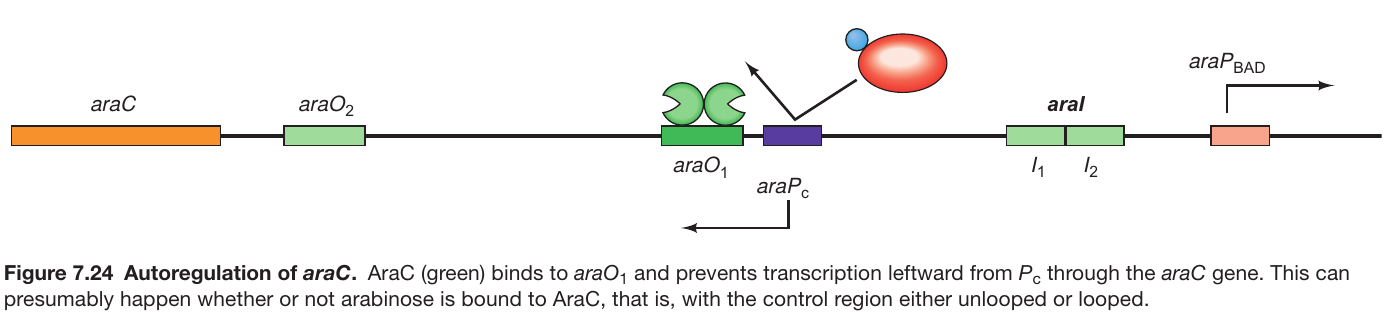
AraC autoregulates its Own Expression
 |
|---|
| Molecular Biology; Weaver, Robert: Fifth edition |
-
Background:
- araO1’s function: Not involved in the repression of araBAD transcription.
- Location: Positioned to control the transcription of the araC gene (refer to Figure 7.24).
-
Autoregulation Mechanism:
- Transcription Direction: The araC gene is transcribed leftward from the Pc promoter.
- Binding to araO1: As AraC levels increase, it binds to araO1.
- Inhibition of Transcription: The binding of AraC to araO1 inhibits further leftward transcription from Pc.
- Control of AraC Levels: The process prevents excessive accumulation of AraC, ensuring self-regulation of its synthesis levels.
-
Conclusion:
- Autoregulation: The mechanism through which AraC controls its own synthesis by inhibiting transcription upon binding to araO1 is termed autoregulation.
- Regulatory Balance: This autoregulation helps maintain a balance in AraC levels, avoiding its overproduction and ensuring optimal functioning of the operon regulatory system.-
Positive promoter elements
Lactose operon promoter:
-35 and -10 sequences are the two binding site of the CRP-cAMP. One of them are mutated, the binding still performed. But simultaneous mutation would decrease the expression level.
- CAP is a universal transcription promoter
- CAP is a sequence-specific DNA binding protein
- CAP responds to cAMP levels in the cell.
- it binds a dimer
- cAMP-CAP binding is required for high expression of the lac operon-- conserves the cells energy.
- TGTGA sequence
So far, hundreds of genes have been discovered which are under the control of the CAP-cAMP complex. Among them are genes involved in cell division, carbon metabolism, flagellar synthesis (controlling the motility), cell energy, nitrogen utilization, iron uptake and drug resistance
CAP recruitment
- Forma- tion of the closed promoter complex
- conversion of the closed promoter complex to the open promoter complex.
P19
DNA binding doesn’t change so much, but the transcription activation is different.
Eukaryol: homodimer
Prokaryotic /heterodimers
Class 1: far away from the gene
Class 2: closer to the gene
Kb creating close
k2: transition close to the open
Different promoter classes Utilize different protein regions.
Molecule and Cellular Biology 4
https://karobben.github.io/2023/08/31/LearnNotes/UIUC-AdMC-4/








