Pseudotime Analysis with Monocle: A Beginner's Guide
Psudotime Analysis
Introduction
In the complex world of cellular dynamics and differentiation, tracking the progression of individual cells can seem like tracking a grain of sand on a beach. Fortunately, pseudotime analysis using tools like Monocle has made it more accessible. But what is pseudotime? And why should we use Monocle for it? Let’s dive in.
What is Pseudotime?
Pseudotime is a computational concept used to order cells based on their progression in a particular process, like differentiation. Rather than relying on actual time (which isn’t available in static single-cell datasets), pseudotime arranges cells in a continuum that reflects their progression state. In simple words, it’s like retracing the journey of a cell from its starting point to its destination, based on markers or genes it expresses.
Why Do We Use Pseudotime Analysis?
Mode details in Monocle
-
Choosing Genes for Trajectory: Monocle selects genes that define cellular progress for its machine learning approach. This feature selection is vital as it impacts the trajectory shape. While some low-expressed genes can be noisy, they might hold essential cellular information. Monocle identifies genes with meaningful variations to structure the data. Users can either let Monocle autonomously choose genes (unsupervised) or input known genes to guide the trajectory (semi-supervised).
-
Dimensionality Reduction: After gene selection, Monocle reduces the data’s dimensionality using the Reversed Graph Embedding algorithm.
-
Pseudotime Cell Ordering: Monocle projects expression data into a reduced dimension space to determine the cellular trajectory. It presumes a tree-structured trajectory with a root and leaves. Monocle’s objective is to best fit this tree to the data. Cells begin at the root and move along the trajectory, making decisions at branches until reaching a leaf. A cell’s pseudotime is the distance from its position back to the root.
How Does Monocle Work?
Monocle stands out in the world of pseudotime analysis because of its robustness and flexibility. Here’s a simplified overview of how it functions:
-
Expression Data Input: Monocle takes in single-cell RNA-sequencing data. Each cell’s gene expression profile serves as a unique identifier of its state.
-
Dimensionality Reduction: The vastness of gene expression data is reduced into manageable dimensions using techniques like DDRTree or UMAP.
-
Trajectory Construction: Using the reduced dimensions, Monocle constructs a trajectory that orders cells based on their progression.
-
Pseudotime Assignment: Cells are then assigned a pseudotime value based on their position in the trajectory.
What Makes Monocle Special?
Several tools offer pseudotime analysis, but Monocle has some distinct advantages:
-
Handling Complex Trajectories: Monocle can discern branched trajectories, which is crucial when cells can differentiate into multiple types.
-
Flexibility: It is amenable to various types of analyses beyond just pseudotime, such as differential expression analysis across trajectories.
Comparison with Other Techniques
-
Monocle vs. Wanderlust: While both are used for trajectory analysis, Wanderlust was primarily designed for mass cytometry data. Monocle offers a more generalized approach, suitable for scRNA-seq data.
-
Monocle vs. Slingshot: Slingshot is another tool for pseudotime analysis of scRNA-seq data. While Slingshot excels in simplicity and user-friendly plotting functions, Monocle’s capability to handle more complex trajectories gives it an edge in certain scenarios.
Pros and Cons of Using Monocle
Advantages:
- Robust in handling complex cellular trajectories.
- Suitable for various analyses.
- Well-documented and supported by an active community.
Disadvantages:
- Might have a steeper learning curve for beginners.
- Certain computations can be time-intensive.
Conclusion
Pseudotime analysis using Monocle offers a deep dive into cellular dynamics, helping researchers unravel mysteries of cell fate decisions, disease progression, and more. While it’s one of many tools available, its capability to deal with complexity makes it a choice worth considering. Like any tool, its efficacy is determined by the skill and knowledge of the user, so if you’re looking to use Monocle, investing time in understanding its nuances will be immensely rewarding.
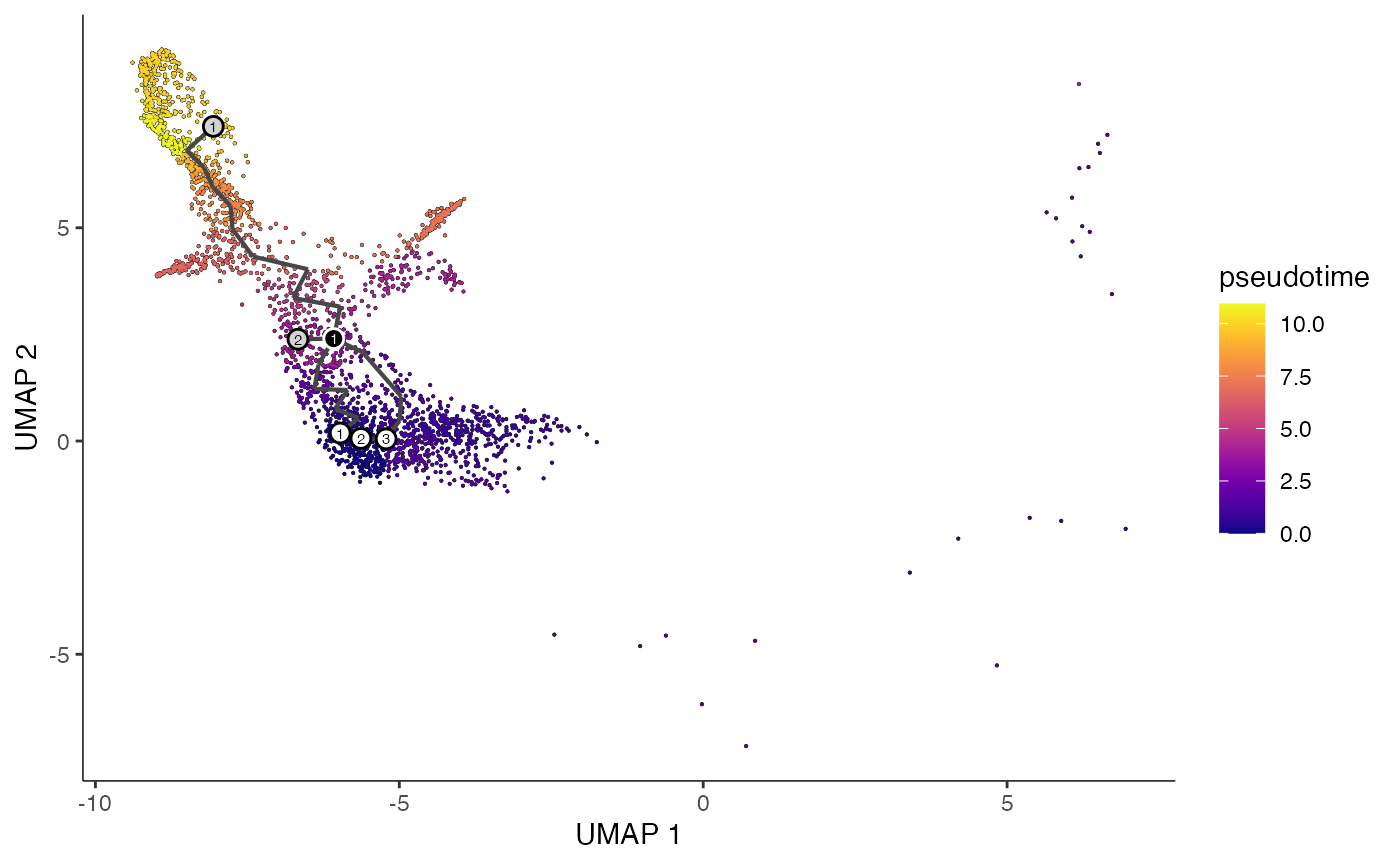
Pepeline from Seurat to Monocle
First, we need to use the SeuratWrappers library to conver the Seurat object into Monocle manageble data[1].
|
 |
|---|
| MonoCle |
Pseudotime Analysis with Monocle: A Beginner's Guide