Molecule and Cellular Biology 6
Points for the lecture:
- λ life cycle is regulated through molecular events
- Events are ‘tuned’ for propagation of the phage
- Competition between factors dictates activity state
λ Phage
How λ Gets In
-
Components and Structure
- Genome: 48,502 bp of double-stranded DNA (dsDNA).
- Chromosome in the capsid:
- Structure: Linear dsDNA.
- Special Feature: 12 nt single-stranded DNA (ssDNA) cohesive termini.
- Capsid:
- Head: Comprises products of B, C, Nu3, D, and E genes.
- Tail: Formed by products of J and H genes.
-
Infection Process
- Attachment: The phage attaches to the host cell’s maltose receptor, a product of the E. coli lamB gene.
- Injection: Injects its linear dsDNA chromosome into the host cell.
- Circularization:
- Mechanism: Annealing occurs between complementary 3’ overhanging cos sites.
- Outcome: Linear dsDNA transforms into a circular form, facilitating the integration into the host’s genome.
-
Notes
- The cohesive termini and the specific structure of the capsid play crucial roles in the bacteriophage’s ability to infect and integrate into the host genome.
- Understanding the precise infection mechanism can offer insights into bacterial immunity and potential applications in bacterial genetics research.
Life Choice Depends on Genetic Switch
1. Lytic Mode
- Initial Phase:
- DNA Entry: Phage DNA enters the host cell.
- Transcription: Host RNA polymerase transcribes phage DNA.
- Translation: Phage mRNAs are translated to produce phage proteins.
- Replication Phase:
- DNA Replication: Phage DNA replicates.
- Assembly: New phages are assembled from DNA and protein components.
- Final Phase:
- Lysis: The host cell undergoes lysis, releasing progeny phages.
- Result: The host cell is killed.
2. Lysogenic Mode
- Initial Phase:
- DNA Entry: Phage DNA enters the cell.
- Early Gene Expression: Early genes are transcribed and translated.
- Repressor Role:
- λ Repressor (CI) Appearance: A 27-kD phage protein appears.
- Binding: CI binds to two phage operator regions.
- Effect: It shuts down transcription of all phage genes except for cI (gene coding for λ repressor).
- Integration Phase:
- Integration: Phage DNA integrates into the host genome.
- Lysogen Formation: The host with integrated phage DNA is termed a lysogen.
- Prophage: The integrated DNA is referred to as a prophage.
- Maintenance Phase:
- Reproduction: The phage DNA replicates alongside host DNA.
- Advantage: Allows the phage to multiply without creating new phage particles, giving it a “free ride.”
- Induction Phase (Conditional):
- Trigger: Exposure to mutagenic chemicals or radiation.
- Effect: The lysogenic cycle can be broken, shifting the phage into the lytic phase.
Notes
- λ Phage Characteristics: It is a temperate phage, meaning it can follow both lytic and lysogenic paths.
- Versatility: λ phage showcases more versatility compared to virulent phages like T2, T4, T7, and SPO1, which always follow the lytic path.
- Lysogenic Stability: The lysogenic state can be stable indefinitely, beneficial for the phage as it ensures its survival and replication without killing the host.
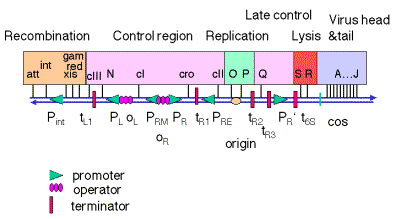
λ Genome is Organized By Function
 |
|---|
| © bx.psu.edu |
-
HEAD & TAIL Region (left side of the RECOMBINATION Region)
- Structure: Encodes for the structural proteins constituting the phage’s capsid.
- Terminase Enzyme: Required for processing rolling circle multimers into unit genome-length pieces during DNA packaging.
-
RECOMBINATION Region
- Genes:
- Int: Necessary for the integration of the phage into the bacterial host chromosome during lysogenic growth.
- Xis: Facilitates the excision of the phage from the host chromosome during induction.
- Others: A variety of other genes facilitating integration and excision processes.
- Genes:
-
REGULATION Region
- Immunity Region: Involved in the phage’s self-immunity processes.
- Switch Control: Houses genes controlling the switch between lysogenic and lytic growth.
- Q Antiterminator Protein: Part of the regulatory mechanism.
- Anti-Q RNA: Works alongside the Q protein in regulation.
- PR’: Constitutes a secondary regulation region, working in conjunction with Q protein and Anti-Q RNA.
-
REPLICATION Region
- Genes:
- O: Replication protein gene.
- P: Another gene involved in replication.
- Origin of Replication: The starting point for the DNA replication process.
- Genes:
-
LYSIS Region
- Gene Count: Contains four genes.
- Function: The genes in this region are generally involved in the lysis of the host cell, facilitating the release of new phage particles.
Note
- Different Regions: Each region in the bacteriophage genome is designated for a different function, playing a crucial role in the phage’s life cycle, either helping in its replication, regulation of its life cycle, or in the processes leading to the integration into or excision from the host genome.
λ Inserts into the Host Genome by Recombination
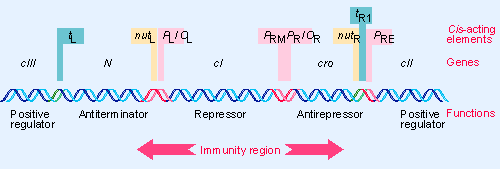
λ has 7 Promoters
- PR expresses the replication genes as well as the anti-repressor, Cro, the transcriptional activator cII, and the anti-terminator, Q protein.
- PL expresses the recombination genes as well as the anti-terminator, N, and the cIII protein.
- PR’ expresses the lysis proteins, and the head and tail proteins.
- PRE expresses the repressor gene, cI, to establish lysogeny.
- PRM expresses the repressor gene, cI, to maintain lysogeny.
- PI expresses the int gene to synthesize the Integrase protein.
- PaQ drives synthesis of a short anti-sense RNA which blocks translation of Q gene mRNA.
Protein Expression is Stage Dependent
| Event | l Gene Expressed | Comment |
|---|---|---|
| Initial infection | Cro, N | Only N and Cro are synthesized until the decision point is reached |
| Lytic pathway | Cro, N, Q, late genes | Cro predominates at operators, N and Q are antiterminators |
| Lysogenic pathway | cI, cII, cIII, int | cII and cIII collaborate to establish cI synthesis; after genome integration, only cI is expressed during maintenance of lysogeny. |
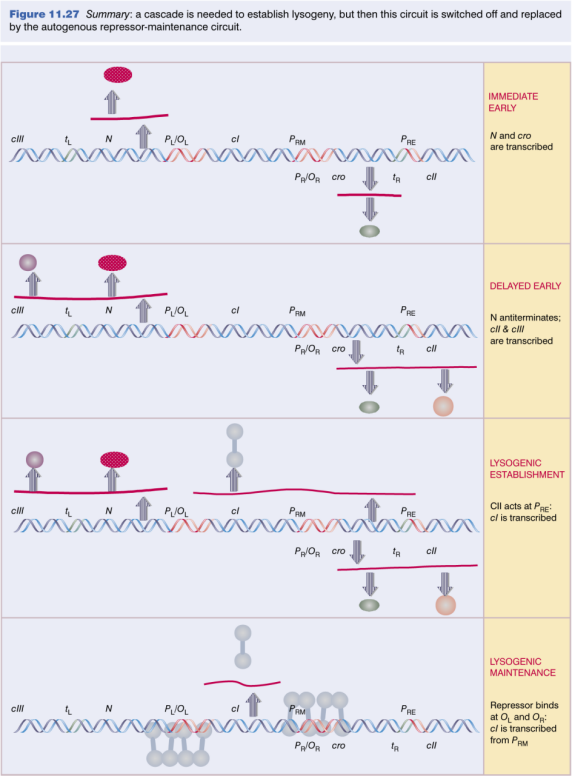
Consequences of Early Gene Expression
- Cro repressor mRNA made from PR and translated.
- N antiterminator mRNA made from PL and translated. Allows transcription and translation of cII and cIII.
- cII/cIII proteins activate PRE, causing transcription of cI
- Concentrations of cI and Cro proteins build up
Initial Events
Molecular Decision-Making: Choice between Lysis & Lysogeny
- Determined by relative concentrations of cI and Cro, which act oppositely on PRM the promoter for Repressor Maintenance
- PRM requires cI protein, therefore not active just after infection. Inhibited by Cro.
- Cro lower affinity for PRM but is made earlier.
- Cro binds same operator sites as cI, but not as efficiently or stably. At low levels, will slow down, but not stop expression of N , cIII (PL) or cII, P, Q (PR). At high levels, cII and cIII are prevented from being transcribed, so no cI.
λ Immunity Region
 |
|---|
| © genes.atspace.org |
Lysogeny
 |
|---|
| Unknown |
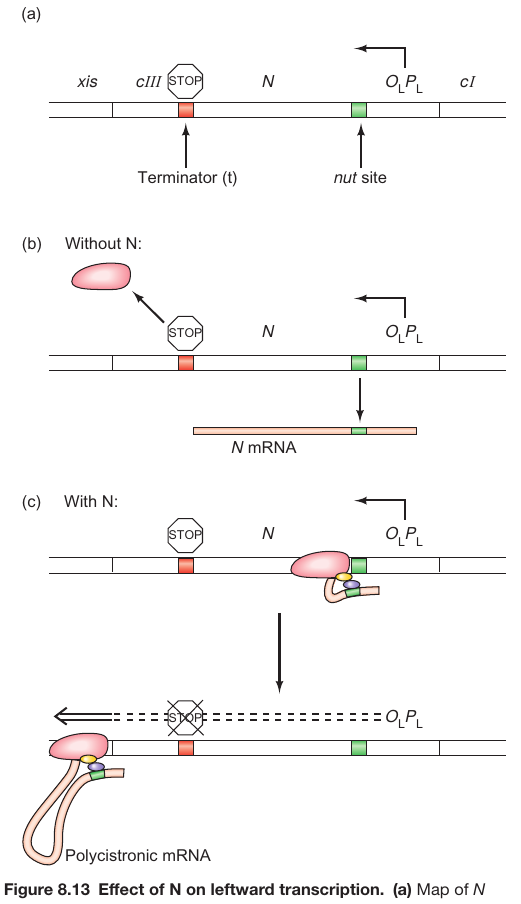
N is an Anti-Terminator
 |
 |
|---|---|
| Molecular Biology; Weaver, Robert: Fifth edition; P206 | P207; Figure 8.14 |
-
Physical and Functional Layout
- Location: Situated downstream of the PL promoter and its respective operator, OL.
- Nut Site: Contains specific sequences, box A and box B, which play crucial roles in the functioning of the N protein.
-
Functionality in the Absence of N Protein
- Transcription Start: Commences at the PL promoter.
- Transcription Termination: Occurs at a terminator site situated downstream of the N gene. The RNA polymerase releases the N mRNA here.
-
Functionality in the Presence of N Protein
- N Protein Synthesis: Once the N gene has been transcribed, the N protein materializes.
- Binding to Nut Site: The N protein binds to the nut site on the transcript.
- Alteration of RNA Polymerase: The N protein collaborates with a complex of host proteins, altering the RNA polymerase to overlook the terminator site and proceed with transcription into the delayed early genes.
-
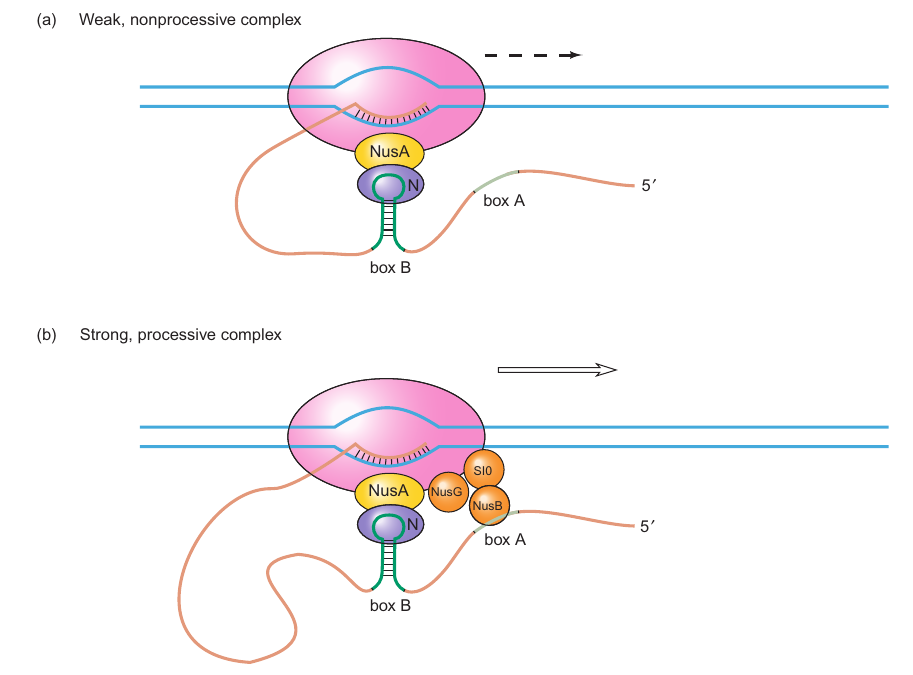
Involvement of Host Proteins
- Nus Proteins: These include NusA, NusB, and NusG, which have roles in both the phage and host cell processes.
- Ribosomal S10 Protein: Participates in protein synthesis in the host cell.
- Antitermination: N and NusA can facilitate antitermination in close proximity to the nut site, forming a short-range antitermination complex with the RNA polymerase.
-
Processive Antitermination
- Long-Range Effect: In natural circumstances, antitermination occurs hundreds of base pairs downstream of nut sites, requiring the involvement of all Nus proteins and S10 for stability.
- Persistent Complex: A stable complex is formed, continuing until reaching the terminator.
-
Nut Site Interaction
- RNA Interaction: Rather than interacting with the nut site itself, the complex interacts with its transcript.
- RNA-binding Domain: The region in N essential for nut recognition features an arginine-rich domain akin to RNA-binding domains.
- Protection from RNase: A full assembly of the five proteins in the complex shields both boxes A and B from RNase attacks.
-
RNA Loop Formation
- Sustained Signal: The RNA between the nut site transcript and the RNA polymerase forms a loop, maintaining the association of N with both, thus providing a continuous signal to the polymerase until it reaches the terminator.
- Evidence: Experiments have indicated that alterations in the RNA polymerase b-subunit gene can obstruct N-mediated antitermination, suggesting a potential association between the RNA polymerase, N, and the nut site transcript during transcription.
-
Conclusion
The gene N encodes for the N protein which plays a pivotal role in the antitermination process during the phage lifecycle. It acts by binding to the nut site of the transcript and altering RNA polymerase activity, facilitating transcription beyond the terminator site. This mechanism leverages both short-range and processive antitermination, guided by a complex interplay of different proteins and the intricate structure of the nut site and its transcript. The stability and sustained activity of this system are central to its function, maintaining a persistent signal through RNA loop formations that guide the RNA polymerase over substantial distances.
N Prevents Stem-loop Formation
 |
|---|
| P208; Figure 8.15 |
-
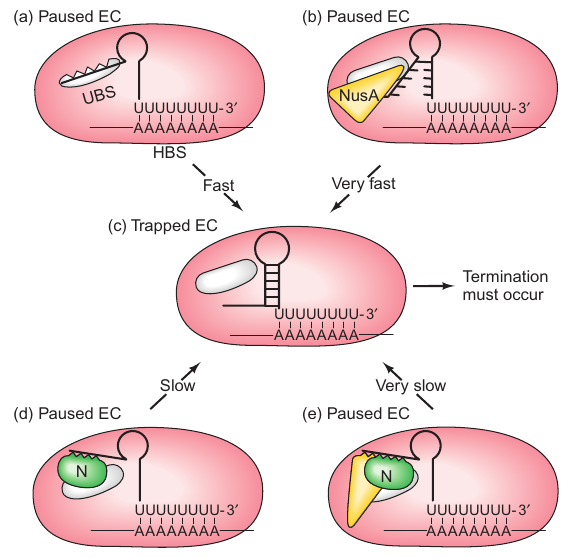
Background
- Initial Hypothesis: N restricts the RNA polymerase pausing vital for termination.
- Research by Gusarov and Nudler (2001): Contradicted the initial hypothesis, highlighting that N doesn’t significantly affect pausing but instead influences the formation of the terminator hairpin.
-
Mechanism
- N Binding to RNA: N binds to the portion of RNA supposed to form the upstream part of the terminator hairpin, thereby slowing down its formation.
- Preventing Hairpin Formation: Without the hairpin structure, the termination process cannot proceed, a mechanism somewhat similar to overriding transcription attenuation seen in the trp operon.
-
Role of NusA
- Interaction with Elongation Complex (EC): As EC synthesizes a string of U’s, it pauses after adding the seventh nucleotide, positioning the potential upstream part of the hairpin to bind to the RNA polymerase’s upstream binding site (UBS).
- Time Constraint: The pause lasts for about 2 seconds, within which the hairpin should form to ensure termination; otherwise, the polymerase progresses without terminating.
- Stimulation of Termination: NusA weakens the connection between the potential upstream part of the hairpin and the UBS, encouraging quick hairpin formation and subsequently promoting termination.
-
Comprehensive Model (Gusarov and Nudler, 2001)
- Dual Binding: Both N and NusA bind to the RNA segment meant to form the upstream part of the hairpin.
- Blocking Hairpin Formation: By binding to the RNA segment, N prevents the quick formation of the hairpin.
- NusA’s Role: While connected to N, NusA also associates with the RNA segment, further slowing down the hairpin formation.
- Outcome: Due to the hindered hairpin formation, RNA polymerase moves forward without engaging in the termination process.
-
Conclusion
N prevents termination not by limiting RNA polymerase pausing but by binding to the RNA segment that is crucial for forming the terminator hairpin, consequently slowing down its formation. NusA plays a dual role, both facilitating and slowing down hairpin formation depending on its interactions with either the RNA or N. The intricate interaction between N, NusA, and the RNA orchestrates whether the termination proceeds or not.
PL
 |
|---|
| source: wiki |
In addition to N, the early transcript of PL codes for:
- cIII
required to protect the activator protein, cII - Xis
normally required for excision of a prophage - Int
normally required for integration of a prophage.
Background
- Context: Early lytic growth phase of bacteriophages.
- Concerned Molecules: Xis and Int proteins.
- Primary Regulator: N protein.
Mechanism
- Role of N Protein:
- Transcriptional Impact: Influences RNA polymerase to bypass the tI transcription terminator and continue transcription through the sib region.
- Attachment: Remains attached to the ternary transcription complex.
- Sib Region:
- Characteristic: Houses the signal for an RNase III processing site.
- Effect on mRNA: Transcripts passing through this region undergo structural changes to form a hairpin configuration.
- Action of RNase III:
- Recognition: Identifies the hairpin structure in the transcripts.
- Cleavage: Splits the transcript at the identified site, leaving free 3’ ends.
- Exonuclease Activity:
- Degradation: Enzymes present in the cell degrade the free 3’ ends further.
- Impact on Xis and Int: The coding regions for Xis and Int largely get destroyed before translation can occur, reducing their expression significantly.
Retroregulation
- Definition: A regulatory mechanism where the stability and translatability of mRNA are controlled post-transcriptionally, affecting the subsequent expression of certain genes (in this case, Xis and Int).
- Outcome in the Context: Ensures limited expression of Xis and Int during the early lytic growth phase, maintaining control over the developmental pathway of the bacteriophage through intricate regulation at the RNA level.
Conclusion
- Efficiency in Regulation: Retroregulation efficiently controls the expression of Xis and Int proteins during early lytic growth through a series of RNA modifications and processing, preventing their significant expression and translation during this phase.
- Regulatory Network: The process involves a network of actions including the N protein’s influence on RNA polymerase, hairpin formation guided by the sib region signal, RNase III-mediated cleavage, and exonuclease-induced degradation, demonstrating a tight regulatory mechanism at work during bacteriophage development.
cIII Inhibits Proteolysis of cII
cII half-life alone is < 1min but with cIII it is ~ 5 min
Molecule and Cellular Biology 6
https://karobben.github.io/2023/09/07/LearnNotes/UIUC-AdMC-6/