require(org.Hs.eg.db)
library(pathview)
data(gse16873.d)
data(demo.paths)
sim.cpd.data=sim.mol.data(mol.type="cpd", nmol=3000)
gse16873.t <- apply(gse16873.d, 1, function(x) t.test(x,
alternative = "two.sided")$p.value)
i <- 3
sim.cpd.data2 = matrix(sample(sim.cpd.data, 18000, replace = T), ncol = 6)
rownames(sim.cpd.data2) = names(sim.cpd.data)
colnames(sim.cpd.data2) = paste("exp", 1:6, sep = "")
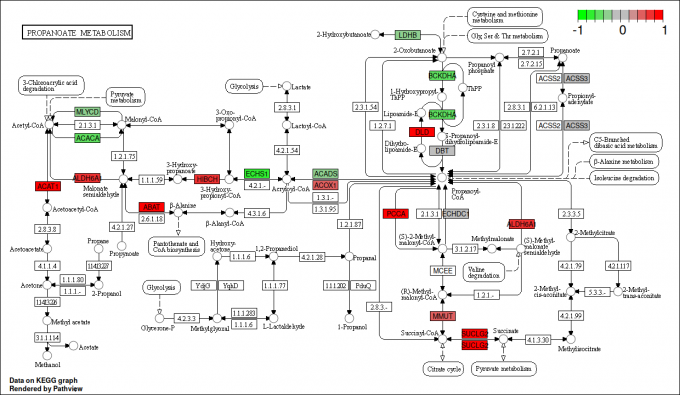
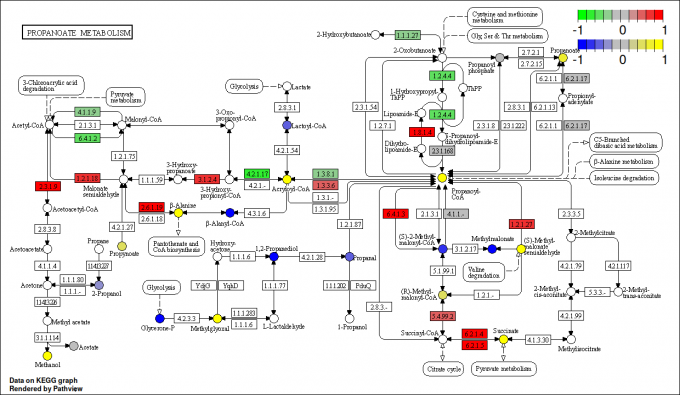
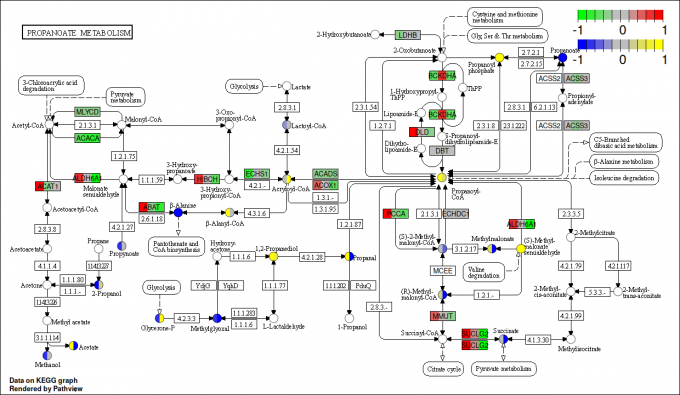
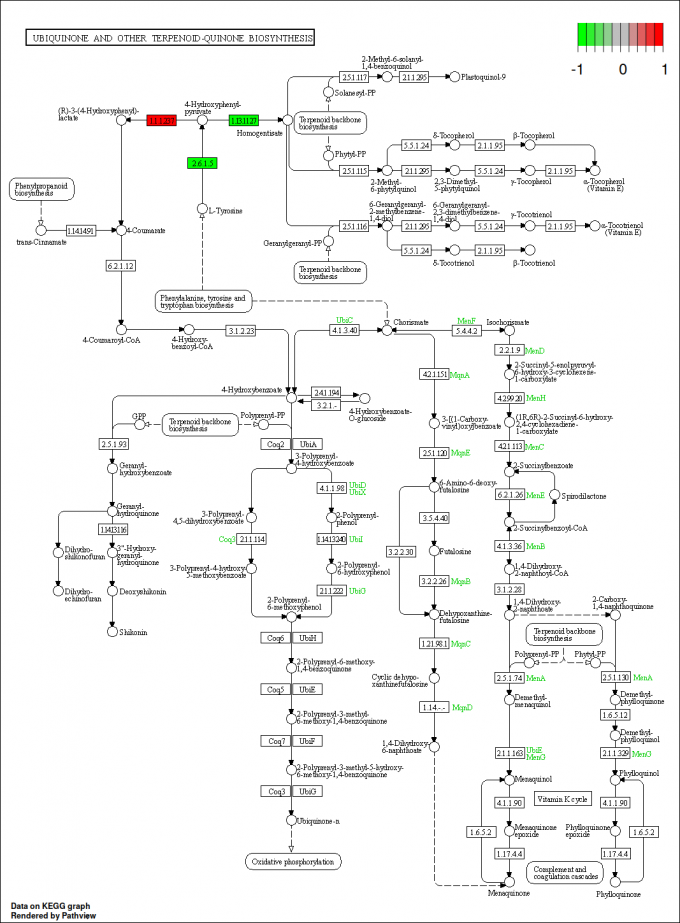
pv.out <- pathview(gene.data = gse16873.d[, 1:3],
cpd.data = sim.cpd.data2[, 1:2], pathway.id = demo.paths$sel.paths[i],
species = "hsa", out.suffix = "gse16873.cpd.3-2s.2layer",
keys.align = "y", kegg.native = T, match.data = F, multi.state = T,
same.layer = F)
|