Fasta Sequences Align
Quick look about the data
Suppose there is a fasta file name as LYSV-NCBI.fasta
Have a quick look
grep ">" LYSV-NCBI.fasta |wc
wc LYSV-NCBI.fasta
echo $(wc LYSV-NCBI.fasta| awk '{print $2}')-$(grep ">" LYSV-NCBI.fasta |wc |awk '{print $2}')|bc
|
124 1114 8363
18250 19116 1282572 LYSV-NCBI.fasta
18002
|
As you can see, there are 124 sequences and roughly about 18002 bases
Let’s have a quick look at the sequences by Seq-view
Seq-view1.3 -i LYSV-NCBI.fasta
|
ClustalW2
Install
url: click me
wget http://www.clustal.org/download/current/clustalw-2.1.tar.gz
tar -zxvf clustalw-2.1.tar.gz
cd clustalw-2.1/
./configure
make
make install
|
clustalw2 -QUICKTREE -OUTPUT=FASTA -INFILE=LYSV-NCBI.fasta
|
It takes less than 20 mins
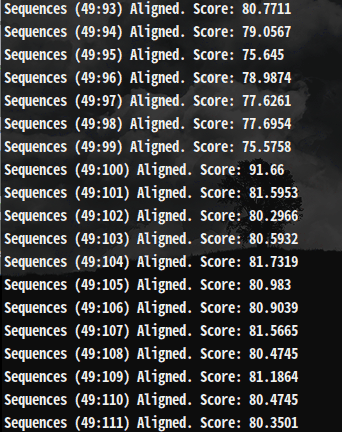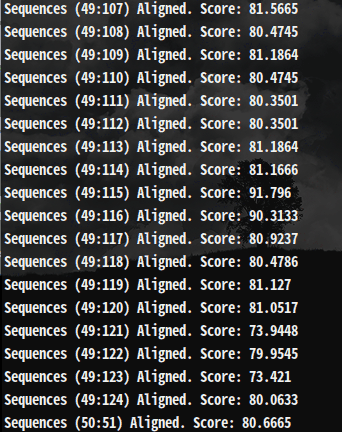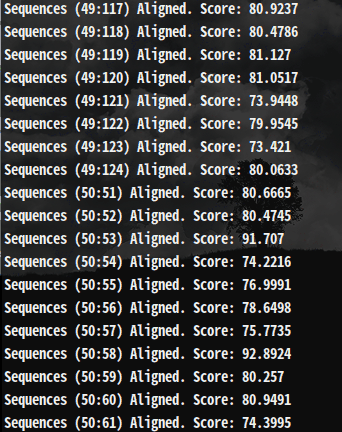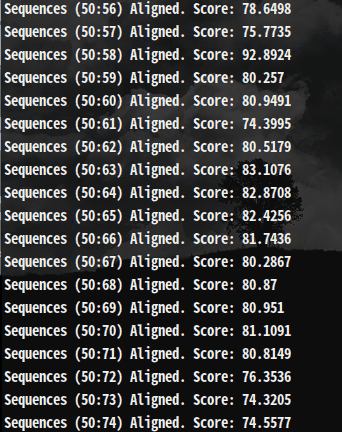
Quick look
Seq-view1.3 -i LYSV-NCBI.fasta -a 90
|
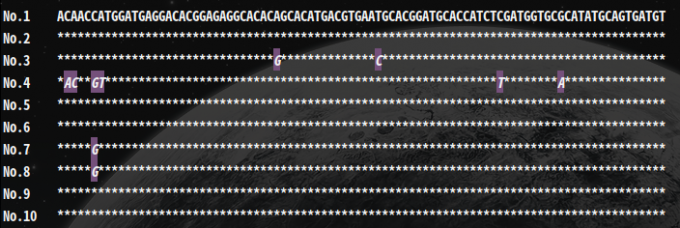
ALign prarmeters
UPGAM Tree
clustalw2 -QUICKTREE -OUTPUT=FASTA -INFILE=LYSV-NCBI.fasta -CLUSTERING=UPGMA -BOOTSTRAP=1000
clustalw2 -QUICKTREE -OUTPUT=FASTA -INFILE=LYSV-NCBI.fasta -CLUSTERING=NJ -BOOTSTRAP=1000
|
PS:
ClustalW well not change the " ’ " from the name of the sequence which may cause trouble in the tree file.
Muscle
apt install muscle
time muscle -in LYSV-NCBI.fasta -out 123.fa
|
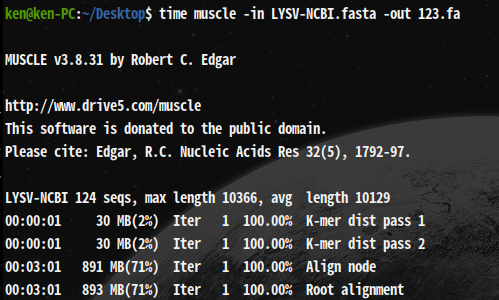
real 17m59.470s
user 17m59.034s
sys 0m0.256s
|
It takes about 18min as well as ClustelW2
Tcoffee
Source: 🏠 T-Coffee
Protein sequences
t_coffee sample_seq1.fasta
t_coffee sample_seq1.fasta -mode quickaln
t_coffee sample_seq1.fasta -mode mcoffee
t_coffee sample_seq1.fasta -mode expresso
t_coffee sample_seq1.fasta -mode psicoffee
t_coffee sample_seq1.fasta -mode accurate
|
DNA sequences
t_coffee sample_dnaseq1.fasta
t_coffee sample_dnaseq1.fasta -mode procoffee
|
RNA sequences
t_coffee sample_rnaseq1.fasta
t_coffee sample_rnaseq1.fasta -mode rcoffee
t_coffee sample_rnaseq1.fasta -mode rcoffee_consan
t_coffee sample_rnaseq1.fasta -mode rmcoffee
|
Full Tutorial: 🏠🏠🏠












