Complicate Nutrition Data Sheet Visualization
Example Data A <- read.table("Digestive_Enzymes.csv",sep=',',header=T)colname and rowname.± and abc followed with the numbers.Github
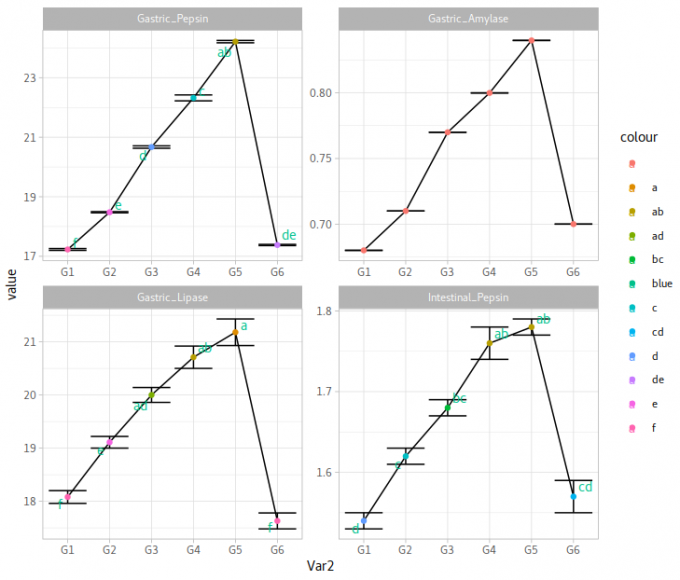
Gastric_Pepsin Gastric_Amylase Gastric_Lipase Intestinal_PepsinG1 17 .23 ±0 .03 f 0 .68 ±0 .00 18 .08 ±0 .12 f 1 .54 ±0 .01 dG2 18 .48 ±0 .02 e 0 .71 ±0 .00 19 .11 ±0 .11 e 1 .62 ±0 .01 cG3 20 .67 ±0 .04 d 0 .77 ±0 .00 20 .0 ±0 .14 ad 1 .68 ±0 .01 bcG4 22 .32 ±0 .10 c 0 .80 ±0 .00 20 .71 ±0 .21 ab 1 .76 ±0 .02 abG5 24 .21 ±0 .04 ab 0 .84 ±0 .00 21 .18 ±0 .25 a 1 .78 ±0 .01 abG6 17 .38 ±0 .02 de 0 .70 ±0 .00 17 .63 ±0 .15 f 1 .57 ±0 .02 cd
Data Clean
NutriSplit <- function (A){c ()c ()for (i in A){as.character (i),'±' ),"[" ,1 )as.character (i),'±' ),"[" ,2 )c (A1,tmp1)c (A2,tmp2)as.numeric (A1)"[^0-9.]" , "" , A2)"[^a-z]" , "" , A2)as.numeric (A2_1),nrow = nrow(A))c ()return (Result)
Plot
library(ggplot2)0.2 )+'blue' ))+ theme_light() +'free' )+45 , hjust=1 ))