import argparse
parser = argparse.ArgumentParser()
parser.add_argument('-i','-I','--input')
parser.add_argument('-o','-O','--output', default="out.png")
args = parser.parse_args()
INPUT = args.input
OUTPUT = args.output
import os
import matplotlib as mpl
import matplotlib.pyplot as plt
from Bio import SeqIO
from collections import defaultdict
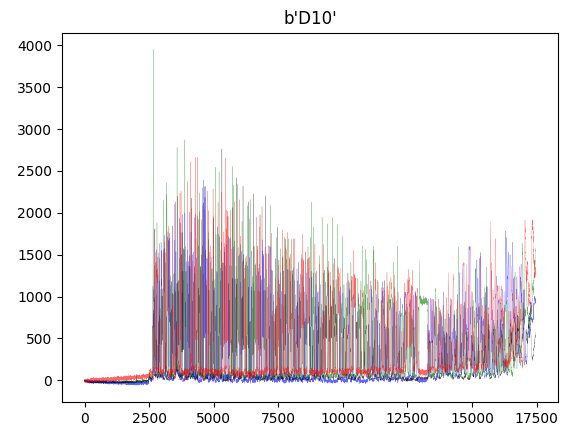
def raw_plot(INPUT):
record = SeqIO.read(INPUT, "abi")
channels = ["DATA1", "DATA2", "DATA3", "DATA4"]
trace = defaultdict(list)
for c in channels:
trace[c] = record.annotations["abif_raw"][c]
plt.plot(trace["DATA2"], color="green" ,alpha=0.6, lw=0.2)
plt.plot(trace["DATA4"], color="blue" ,alpha=0.6, lw=0.2)
plt.plot(trace["DATA1"], color="black" ,alpha=0.6, lw=0.2)
plt.plot(trace["DATA3"], color="red" ,alpha=0.6, lw=0.2)
plt.title(record.annotations['abif_raw']['TUBE1'])
Cmd = "ls "+ str(INPUT)
LIST = os.popen(Cmd).read().split("\n")[:-1]
print(LIST)
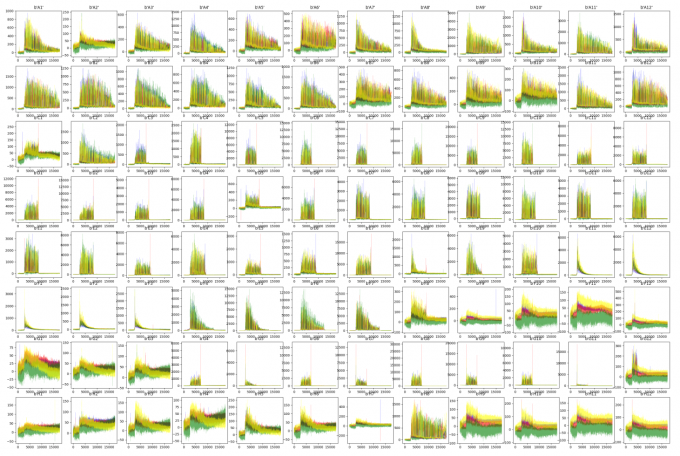
plt.figure(figsize=(14*3, 8*3))
plt.ion()
for i in range(96):
plt.subplot(8,12,i+1)
abi = INPUT+"/"+LIST[i]
print(abi)
raw_plot(abi)
plt.show()
plt.savefig(OUTPUT)
|