用biopython抽取特定長度的序列
1. fasta 讀取和長度統計
1.1 統計
from Bio import SeqIO
Seq = '/media/ken/Data/Yan/RNA-seq/report/2.assembly/out.fa'
Seq = SeqIO.parse(Seq, "fasta")
Len_Seq = []
for seq_record in Seq:
Num = len(str(seq_record.seq))
Len_Seq += [Num]
|
1.2 統計圖
import matplotlib.pyplot as plt
plt.ion()
plt.show()
plt.figure(figsize=(7, 4))
plt.hist(Len_Seq, label=['1st', '2nd'], bins=900)
plt.grid(True)
plt.legend(loc=0)
plt.xlim(xmax=1000,xmin=0)
plt.xlabel('value')
plt.ylabel('frequency')
plt.title('Histogram')
|
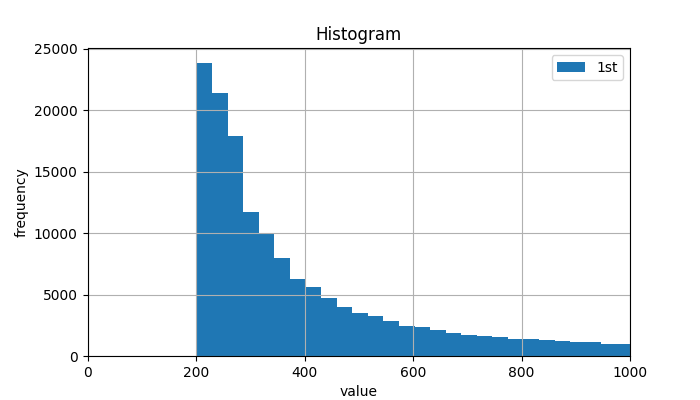
ummm, 決定篩掉30000bp一下的reads
from Bio import SeqIO
Seq = '/media/ken/Data/Yan/RNA-seq/report/2.assembly/out.fa'
Seq = SeqIO.parse(Seq, "fasta")
Num = 4000
Result = []
for seq_record in Seq:
if len(str(seq_record.seq)) > Num:
Result +=[">"+seq_record.id]
Result +=[str(seq_record.seq)]
Fasta = "\n".join(Result)
F = open("out.fa",'w')
F.write(Fasta)
F.close()
|

