Principles of Biochemistry 13 |Glycolysis in Red Blood Cell| Class Notes |HarvardX
Red Blood Cell
 |
|---|
| © Philip W Kuchel, et al. |
Physiological Adaptation of RBC:
- one third of the volume is occupied by hemoglobin.
- lack of intercellular organelles, like Mitochondrial
- allows deformation for moving through narrow capillaries
- lactate fermentation
- Cori cycle: lactate catabolism
Cori Cycle
Rapoport-Luebering shunt
It is a pathway that converts 1, 3-bisphosphoglycerate, one intermediate of glycolysis, into its isomer, 2,3-BPG.
As a bypass pathway, the ATP generation was avoid. As a result, most cells have a very low lever of the 2,3-BPG. But it is very high in RBC since this molecule has a very important function in release of Oxygen.
NADH was produced in Glycolysis works for maintaining the iron in Fe2+ state, which was used to carry the Oxygen.
NADH maintains reduced iron
HMP shut
HMP shut could protect RBC from reactive oxygen species.
Defense against ROS
Oxygen Tranportaion
Conformation Chage:
- R-State (oxygenated, high affinity)
- T-State (non-oxygenated, low affinity)
Affinity
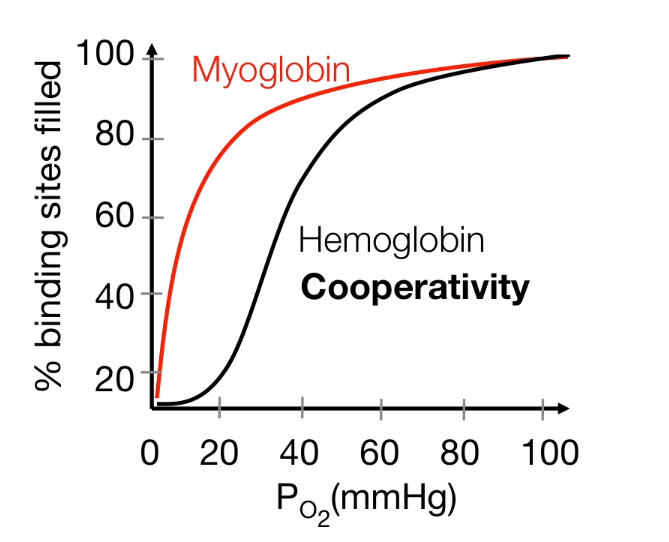
Cooperative binding: Dynamic Oxygen biding
- Releasing about 25% of oxygen
- When it is needed, it could releasing 75% of O2
$$
Hb \underset{O_ 2}{\overset{K1}{\rightleftharpoons}}
HbO_ 2 \underset{O_ 2}{\overset{K2}{\rightleftharpoons}}
Hb(O_ 2)_ 2 \underset{O_ 2}{\overset{K3}{\rightleftharpoons}}
Hb(O_ 2)_ 3 \underset{O_ 2}{\overset{K4}{\rightleftharpoons}}
Hb(O_ 2)_ 4
$$
 |
|---|
| © HarvardX |
Binding affinity change
- Leftward shift: higher affinity
- Rightward shift: lower affinity
PH
The PH changed, which also connected the change of concentration’s level of
CO2:
Muscle: CO3, reducing the PH, which causing Rightward shift, and decreasing the affinity, increasing the releasing of the O2
Lungs: CO3 reduced, PH increased.
2,3-BPG
In the T-shape it was bond
In the R-shape, the bond was crushed and 2.3-BPG was released.
Principles of Biochemistry 13 |Glycolysis in Red Blood Cell| Class Notes |HarvardX
https://karobben.github.io/2021/04/24/LearnNotes/edx-biochm-13/









