Protein Structure|Graduate Biochemistry 3| Tulane
Overview
- What is a protein?
- Folding is chain collapse
- Visualizing structures
- Primary, secondary, tertiary, quaternary structure
- Anfinsen-Merrifield experiments
- Evolution and sequence conservation
- Protein structural motifs
- The Greek Key and β-sandwich
- The coiled-coil
- The EF hand
- Domain structure of large proteins
- Quaternary structure of proteins
Protein Structure-Chain Collapse
A protein of 400 amino acids
| Unfolded | ~500 Å mean diameter fexible polymer chain |
| Folded | ~40 Å diameter; Density ~1.3 g/ml |
PS:
- organic liquids: 0.8-1.0 g/ml
- organic solids: 1.0-1.4 g/ml
Properties of folded proteins
- Folded proteins are as tightly packed as crystals of amino acids
- Secondary structure is an unavoidable consequence of packing
- Most hydrogen bonding groups inside are hydrogen bonded
Visualization of Protein Structure
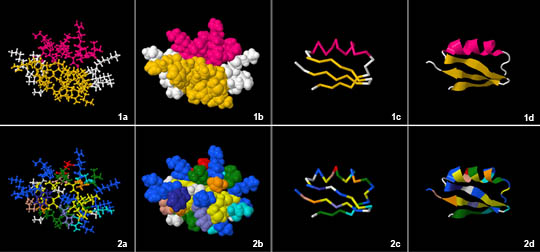 |
|---|
| © asu.edu |
a: NA; b: CPK space-filling; c: Cα trace; d: Ribbon diagram;
Schematic is not show here.
Protein Folding - Levels of Structure
Primary -> Secondary -> Tertiary -> Quaternary
Globular Protein: typically have a fixed (small to moderate) number of subunits, ratio of length to width is small.
Fibrous proteins: large polymeric, ratio of length to width is large
Protein Folding - Levels of Structure
Ribonuclease A:
- 124 residues
- 4 disulfide bonds
- Unfolded with denaturants (urea or guanidine)
The Anfinsen/Merrifield Experiments
More details above: Karobben, Biochemistry_4
- Christian Anfinsen and Robert B. Merrifield
The hypothesis:
All of the information needed to determine the structure of a protein is contained within its amino acid sequence
The experiment:
- Ribonuclease in an enzyme with 4 Cys-Cys disulfide bonds
- The correct linkage is essential for enzymatic activity
- Anfinsen purified ribonuclease from cow pancreas, and then reduced the disulfide bonds with mercaptoethanol and unfolded the protein with 8 Molar urea (It was completely inactive)
- The reduced and denatured protein was put into a solution that favored oxidation of disulfide bonds.
The observations:
- There are 105 different arrangements of the S-S bonds but only the single native arrangement gives active enzyme
- Ribonuclease reoxidized in the presence of 8M Urea gave 1% activity
- Ribonuclease oxidized in the absence of Urea gave ~100% activity
The conclusion:
All of the information needed to determine the structure of a protein is
contained within its amino acid sequence
The confirmation
Robert Merrifield synthesized ribonuclease chemically and got active enzyme. This experiment demonstrated that no cellular machinery or structural “memory” was involved.
Cytochrome c Sequence Alignments
 |
|---|
| © Gulnaz Afzal, et al. |
Sequence conservation in a small, critically important Protein cytochrome c
Substitutions of amino acids can be:
Conservative (similar physical properties. charge etc)
Non conservative (dissimilar physical properties)Phylogenetic Tree of
The degree of sequence relatedness is correlated with the degree
of evolutionary relatedness.
A protein with a conserved sequence will have a conserved structure.
Protein Folding – Structure Motifs
The atomic level 3-dimensional structure is known for ~19,000 proteins
A few hundred folding motifs are adequate to describe most structures
A structural “motif” is a unit of secondary structure topology
Examples of secondary structure motifs
a) βαβ
b) β-hairpins
c) αα
d) β-barrel
e) β-barrel
f) α/β-barrelExamples of structural motifs
TIM-barrel of triosephosphate isomerase
Motif: (βαβα)4
Small β-sheet motifs
A “Greek Key” pattern
- The two Greek key topologies For a 4 stranded β-sheet
- Topology of a 4 stranded β-meander
What 4-stranded β-sheet topologies are found in proteins?
- 24 possible topologies exist
- only 8 have been observed
- the 2 Greek keys and β-meander (above) account for almost 70% of the observed topologies
β sandwich motif
A β-sandwich motif has two beta sheets (often Greek keys) packed face to face.
It’s a small common folding motif.
Hydrophobic α-helix core was been enveloped in the center of the protein.
| © Molview; The crystal structure of APRIL bound to BCMA; PDBID=1XU2; hemoglobin |
Coiled Coils – α-helical bundles
- Dimeric helical pairs have a heptad (7 amino acid) repeat of interacting amino acids.
- Coiled coils form the basis for many long fibrous proteins
Heptad repeat pattern of dimeric
Coiled-coils:
Residues d and a are hydrophobic
Residues g and e form charge pairs
Residues c, b and f are variable
A coiled-coil protein can have from several to hundreds of homologous heptad repeats.
When the ‘d’ resides are leucine/isoleucine the motif is called a “leucine zipper”
Protein Folding – Metal Binding Motif
EF-hand or Helix-loop-Helix motif
 |
|---|
| © Paul T Wilder, et al. |
EF-hand Sequence:
| ------------------------------------Helix | Loop | Helix------------------------------------ |
|---|---|---|
| VkkAFaiID | qDkSGfIEedE | LklFLqnF |
(Parvalbumin β)
lower case – variable
UPPER CASE – conserved
Underlined – involved hydrophobic interaction between helices
Bold – involved in calcium binding
Essential loop Glycine
Target: Calcium atom
[Helix] - [Loop] - [Helix]
- lower case – variable
- UPPER CASE – conserved
- Underlined – involved hydrophobic interaction between helices
- Bold – involved in calcium binding
Protein Architecture - Independent Domains
- Proteins longer than about 200 residues tend to form globular domains
- Domains tend to fold independently and have conserved activities
- A few domain types can make many proteins
- Number of domain types appears limited to a few hundred
An Example of Domains
Proteases in the blood coagulation cascade are good examples of complex multi-domain proteins
- Calcium binding domain
- Kringle domain
- Has a specific pattern of cysteine residues in a span of ~85 residues
- Domains that resemble the epidermal growth factor EGFL domain
- Serine protease domain
Quaternary Protein Structure
- Many proteins are multimeric: Containing more than one polypeptide chain, generally held together by noncovalent interactions
- Homomultimers – composed of copies of the same polypeptide chain
- Heteromultimers – composed of more than one polypeptide chain
- The interactions that drive quaternary structure are the same as those that drive folding
- interaction strengths can range from weak and transient to strong and essential for structure
Aspartate Transcarbamoylase:
- 6 catalytic (C3)2 and 6 regulatory (R2)3 subunits
Protein Structure|Graduate Biochemistry 3| Tulane
https://karobben.github.io/2021/09/16/LearnNotes/tulane-biochem-3/









