Protein Function|Graduate Biochemistry 4| Tulane
Protein Database Bank (PDB)
| © Molview; PDBID=1bkv; Collagen |
Structure and Function of Collagen
- extracellular proteins that provide rigidity and strength to many tissues
- Large bundles of triple helices
- Gly-X-Y
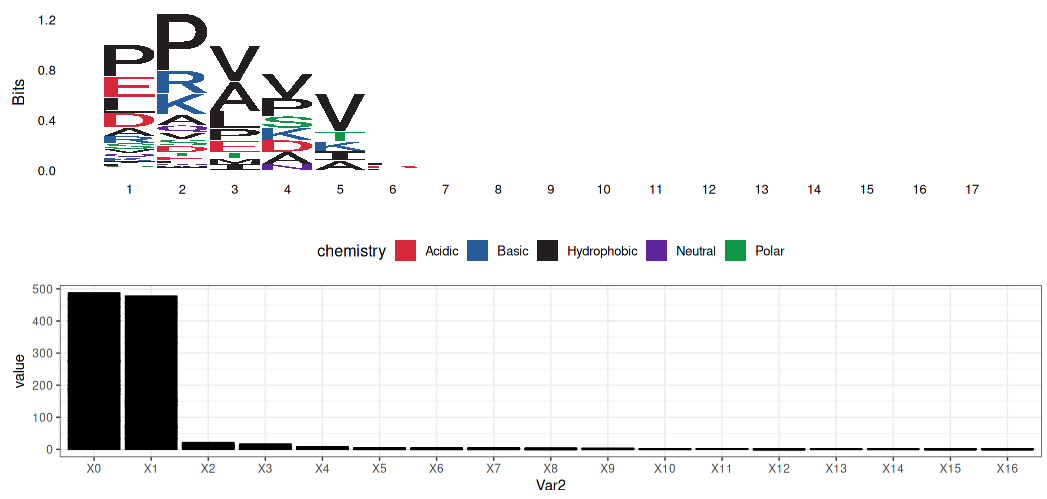
Alanine - Glycine substitute: A is hydrophobically contributed to α-helix composed.
Hydroxyproline…
- stabilized by cross-helix backbone hydrogen bonds
- large bundles of triple helices: A right-handed trimer of left-handed helices.
- Gly-X-Y Pattern: X is mostly proline and many of the Y is hydroxyproline (Gly-Pro-Hyp)
Glycine:
- No side chain: Closer the α-helix
- Hydrophobic: Folding to the center of the helix by hydrophobic effect
Alanine (replacing to Glycine):
- larger than Glycine but could still work similarly to Glycine
Prolein $ Hydroxyproline:
- pull out of the strand
- hydroxyproline work with water
Proline hydrolyzation was carried by Proline hydroxylase and required Vc as co-factor.
$$
Proline + 2-Oxoglutarate + O_ 2 + Fe^ {2+}\overset{Proline hydroxylase}{\longrightarrow} Hydroxyproline + Succinate + CO_ 2 + Fe^ {3+}
$$
$$
V_ c + Fe^ {3+} \to Dehydroascorbate + Fe^ {2+}
$$
- Collagen is stabilized by cross-helix backbone hydrogen bonds
- Gly-NH::O=C-Pro
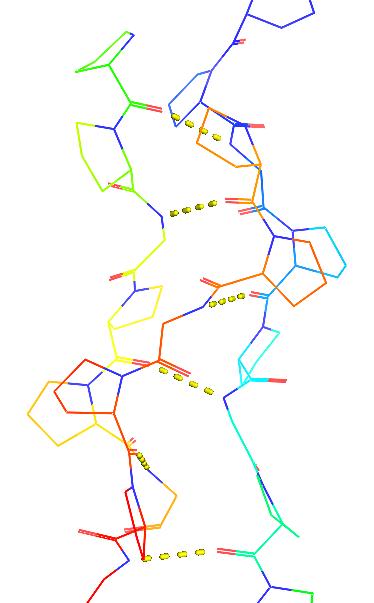 |
|---|
PS: Proline is often disturbing the structure of the α-helix. But why is proline’s favorite in the triple α-helix? (3-D structure)
Globin
Heme
- the oxygen binding cofactor
- Contains a reduced (ferrous, Fe2+) Iron atom.
- The porphyrin ring contains 4 pyrrole groups (A-D)
- Fe2+ has 6 coordination sites.
- 4 pyrrole nitrogens
- The proximal histidine that transmits ligand binding-induced conformational changes to the protein.
- The sixth coordination site that binds various ligands O2, CO, CO2, CN
- A “distal” histidine hydrogen-bonds to the heme-bound O2
Structure and function of myoglobin
- muscle protein that binds O2
- First protein to have its 3D structure determined
- contains a single heme group
| © Molview; PDBID=1MBN; myoglobin |
- First protein to have its 3D structure determined (by John Kendrew, 1956)
- 153 residues long
- 75% α-helical. No β-sheets.
- There are 8 helices designated A-H
| © Molview; PDBID=4hhb; hemoglobin |
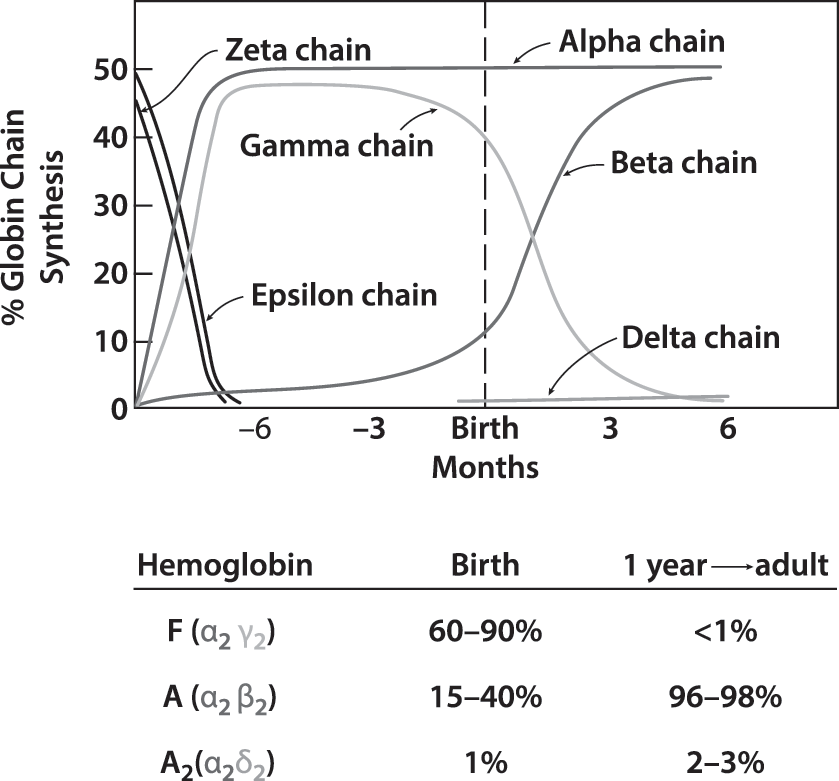
- α2β2 tetramer.
- Human: α, β, γ, δ
- Consist to α2X2(X= β, γ or δ)
- β in adult; and γ in fetal.
 |
|---|
| © Cambridge University |
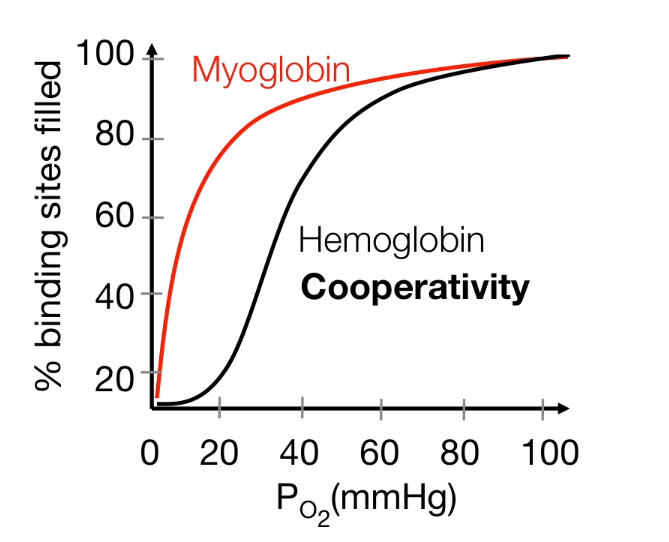
Myoglobin (Mb) versus hemoglobin (Hb)
 |
|---|
| © Jay F Storz, 2002 |
- Hb: sigmoidal (26 torr).
- Mb: hyperbolic (2.8 torr).
- Mb has a greater affinity than Hb for oxygen at all oxygen pressures.
- In the lungs, Hb is saturated
- In tissues, oxygen is released from Hb and transferred to Mb .
 |
|---|
| © HarvardX |
More information about Oxygen Transport: Karobben, Principal of Biochemistry
The Bohr (pH) Effect
Hemoglobin’s oxygen affinity decreases with decreasing pH and increasing CO2, causing the release of oxygen in tissues
PS: Bohr Effect is based on pH effect; pH Changes the conformation of the Myoglobin and Hemoglobin. (R state (oxy-form) to T state (deoxy-form))
- Protonatable His146 of the β subunit is the key of Bohr effect.
- In the tissues (a low pH), His146 forms a salt-bridge with Asp94 and its backbone with Lys40 of the, stabilizing the T state in tissues (Deoxy-form).
- In the lungs (a high pH), the salt-bridge is disrupted, promoting the T to R transition (Oxy-form).
H146 of β interactions in T and R states
α2Lys40::β1His146::β1Asp94
© http://biomodel.uah.es
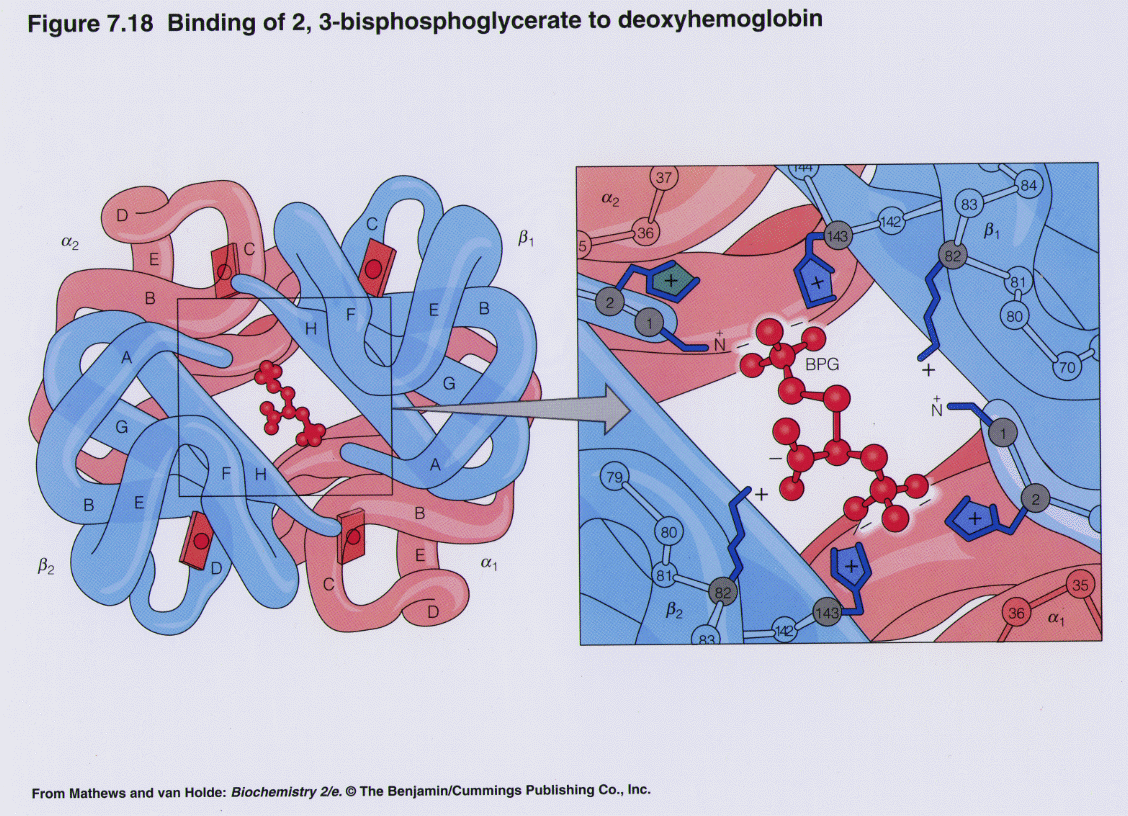
BPG effect:
| © Molview; PDBID=1B86; BPG and Hemoglobin |
- 2,3-bisphosphoglycerate (BPG) binds to hemoglobin (T state) and decreases oxygen affinity.
- BPG is produced within erythrocytes
- BPG is involved in high altitude adaptation
- BPG binds at the interface of β1 and β2 chains stabilized by numerous ionic and hydrogen bonding interactions
- Fetal hemoglobin (γ) doesn’t bind BPG
- b/c His143 -> Ser mutation at the BPG binding site
 |
|---|
| © www3.nd.edu |
Structure and Function of Antibody
- Antibodies function by interacting with foreign (antigen) molecules
- The antigen binding sites are produced by the variable domains from heavy and light chains.
- Several “hypervariable” loops are at the end of the variable domain.
- The two light chains and the two heavy chains are identical: the two antigen-binding sites are identical.
- Antibodies have H2L2 stoichiometry
- H is a “heavy” chain and L is a “light” chain
- Each chain has one variable domain
- All domains (4 in each heavy chain and 2 in each light chains) have the same “immunoglobulin fold”
- α β-sandwich with α 4-stranded and α 3-stranded sheet connected by a disulfide bond.
 |
|---|
| © nthu.edu |
Antibody Peptide Interactions
- Strong and highly specific binding requires numerous favorable interactions between antibody and antigen.
- Involving hydrophobic, ionic and hydrogen bonding interactions.
Myosin
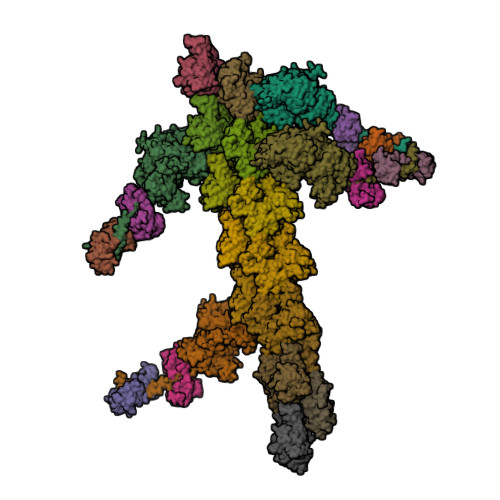 |
|---|
| © PDB; PDBID=1m8q; Myosin |
- Sliding filament mechanism of muscle contraction:
- During contraction, the thin and thick filaments move with respect to one another
- Two heavy chains and several light chains
- Heavy chains are rod-like molecules
- The coiled coil tail domain
- The ATPase head domain
- Arranged with tails overlapping and heads directed toward either end
| Type | Images | Copyright | More |
|---|---|---|---|
| Actin |  |
© PDB, ID=4pkg | - Long filamentous polymer consisting of two strands of globular monomers |
| Tropomyosin |  |
© PDB, ID=1ic2 | - A long, thin two-stranded α-helical rod - In relaxed muscle (low calcium), tropomyosin prevents myosin head from binding to actin |
| Troponin (three subunits) | 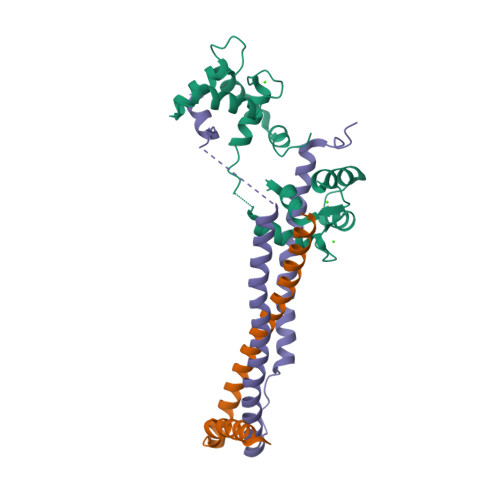 |
© PDB, ID=1j1d | - Tn-I inhibits the actin-myosin interaction - Tn-T binds to tropomyosin - Tn-C binds calcium ions |
Myosin Conformational Changes
 |
|---|
| © Takashi, et al. |
Structural states of myosin during the contractile cycle.
- Without bound nucleotide, myosin is strongly bound to actin (rigor state).
- ATP binding to myosin dissociates the actin-myosin complex.
- ATP is then hydrolyzed in ADP+Pi. There is a swing of the lever arm (green arrow).
- Myosin can rebind to actin
- Release its hydrolysis products and produce its force (green arrow) and again strongly bound to actin without nucleotide bound (power stroke, red arrow)
Protein Function|Graduate Biochemistry 4| Tulane
https://karobben.github.io/2021/09/17/LearnNotes/tulane-biochem-4/









