Lipids, Membranes and Membrane Proteins|Graduate Biochemistry 7| Tulane
Overview
- A cell is a membrane-rich environment
- Classes of lipids
- Structure of lipids
- Polymorphic phase behavior of lipids
- The lipid bilayer membrane
- The fluid mosaic model of biological membranes
- Classes and functions of membrane proteins
- Folding and structure of membrane proteins
- Hydropathy plots
A cell is a membrane-rich environment
Classes of Lipids
- Fatty acid (e.g. palmitate, oleate)
Energy storage metabolite
Building block of other lipids- Triacylglycerols (fat)
- Energy storage
- Glycerophospholipids
Form lipid bilayer membrane
Signal transduction- Sphingolipids
Form lipid bilayer membranes
Signal transduction- Steroids
Rigidify membranes (Cholesterol)
Membrane-crossing hormones (e.g. Estrogen)
Fatty Acids - structure and nomenclature
Fatty acid rules of nomenclature
- Carbon numbering start at carboxyl carbon
- Double bonds are numbered at the first carbon
- cis configuration is normal (trans is specified)
Syntax for nomenclature
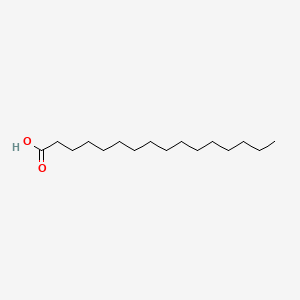
length:#double bonds (list of double bond locations)16:0 = palmitic acid
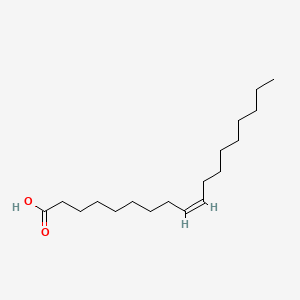
18:1(9) = oleic acid (sometimes written 18:1(Δ9))
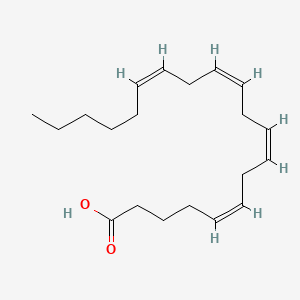
20:4(5,8,11,14) = arachidonic acid
| palmitic acid | oleic acid | arachidonic acid |
|---|---|---|
 |
 |
 |
| © PubChem ID= 985, 445639, 444899 |
Typical eukaryotic fatty acid structure
- Even number of carbons
- Non-conjugated double bonds (double bonds are separated by at least one CH2)
Double bonds are normally cis If a double
bond is trans it is specified in the name
PS:
Even number: Synthesis and degraded of the fatty acid are occur two carbons at a time.
Conjugated double bounds: Conjugated double bounds could make fluorescence. Exp: Retinol, or vitamin A, has five conjugated double bonds and absorbs the violet part of the spectrum, thus appearing as yellow.[1]
Cis: As the picture above, the hydrogen atoms are on the same side.
Palmitic acid
Oleic acid
Classes of diacyl lipids
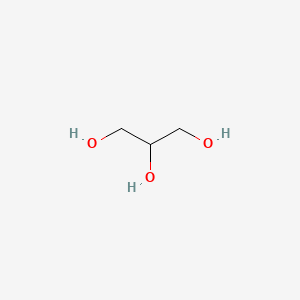
The two backbone molecules in diacyl lipids:
- Glycerol (Glycerophospholipids)
- Serine (Sphingolipids)
Diacyl lipids are the main structural component of bilayers and are the ONLY component that will form bilayers in pure form.
Be able to draw the base of this two lipids.
| Glycerol | Serine |
|---|---|
 |
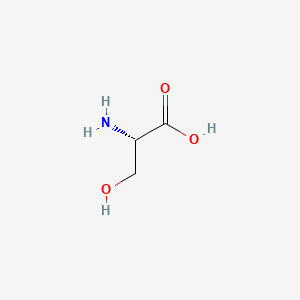 |
| Two fatty acids connect to two hydroxyl groups independently, the rest hydroxyl group connect to the phosphate acid which could connect the variety head. | Two fatty acids connect to Amino group and carboxyl group. R chain connect variety heads |
 |
 |
© PubChem
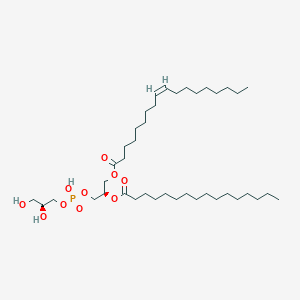
Variety of Glycerophospholipid (head)
 |
 |
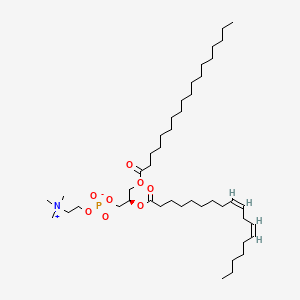 |
| Phosphatidic acid | Phosphatidylethanolamine | Phosphatidylcholine |
 |
 |
 |
| Phosphatidylglycerol | Phosphatidylserine | Phosphatidylinositol |
| © PmdChem |
- second messenger molecule
- Often phosphorylated on one o
several hydroxyls
No head
 |
|---|
| © PubChem ID=6026790 |
A signaling molecule
Phosphatidic acid: is rare in memebrane but it’s intermediate of the metabolic.
Phosphatidylcholine: Most abundant glycerophospholipid found in eukaryotic membrane. Innert, not strong interaction with others. (Outside of the membrane)
Phosphatidylethanolamine: Charge and much activity than Phosphatidylcholine, inside the cell.
Phosphatidylglycerol: also abundant in prokaryotic cell.
Phosphatidylserine: active, inside the cell. Common in eukaryotic cell. It’s not a sugar group, it is a sample carbon base ring.
Phosphatidylinositol: Negative Charged, signaling molecule. Second messenger.
Sphingolipid Headgroups
Sphingomyelin
 |
|
|---|---|
| Sphingomyelin | Phosphocholine group |
Cerebrosides
- Simple sugars
- Galactose, lactose, glucose (uncharged)
Gangliosides
- More complex carbohydrates
(anionic) Found in brain
Fatty acid melting points
The longer the chain, the higher the melting point.
The more double bounds, the lower the melting point.
 |
|---|
| © PubChem ID= 3018404 |
| Melting point:82-95 °C. |
 |
|---|
| © PubChem ID= 5280450 |
| Melting point:-9 °C. |
PS: Cells control the fluid of the membrane by change the type of lipid (melting point) to achieve specific biology purpose.
Four common fatty acid you should remember
| Symbol | Common name | Systematic name | Structure | mp (°C) |
|---|---|---|---|---|
| 14:0 | Myristic Acid | Tetradecanoic acid | CH₃(CH₂)₁₂COOH | 52 |
| 16:0 | Palmitic acid | Hexadecanoic acid | CH₃(CH₂)₁₄COOH | 63.1 |
| 20:0 | Arachidic acid | Eicosanoic acid | CH₃(CH₂)₁₈COOH | 75.4 |
| 24:0 | Lignoceric acid | Tetracosanoic acid | CH₃(CH₂)₂₂COOH | 84.2 |
Phospholipases and cyclooxygenases
 |
|---|
| © Konstantin Balashev |
Phospholipases A1, A2, B, C, and D.
Most phospholipids was cleaved by Phospholipase A2
cyclooxygenases
 |
|---|
| © NV Chandrasekharan |
Cyclooxygenases (COXs) catalyze the rate-limiting step in the production of prostaglandins, which is an signal molecule and both soluble in water and lipids.
Cholesterol and other steroids
 |
|---|
| © PubChem ID= 5997 |
- Cholesterol is a component of biological membranes
- Up to 50% of a membrane’s lipid can be cholesterol
- It rigidifies or stiffens bilayers
- Cholesterol is converted to bile acids (used in digestion) and steroid hormones (used for signaling)
- Steroid hormones freely cross membranes and act directly on nuclear receptors
The hydroxyl group of the cholesterol makes it float on the cell membrane.
In the RBC, the cholesterol in cytomembrane could up to 50% and which makes the membrane very rigidity. (try to keep it’s shape when it through the catepillars) Lake of most membrane proteins. As a contrast, the mitochondria’s membrane does not have cholesterol and has very high fluidity which needs to go constant conformation change. One of the fluidity membrane we know are rod and cone cell which are light recept cells. They need to interact quickly. As a result, they are made highly portion of the saturated fatty acids and very little of cholesterol.
| Steroid Hormones | © PubChem |
|---|---|
 |
 |
 |
 |
These hormones are both soluble in water and lipids. So they can pass membrane easily.
Polymorphic Phase Behavior of Lipids
- All lipids spontaneously separate from water
- The form of the lipid phase depends on the molecular “shape” of the molecule
Head group end
Molecular shap hypothesis:
| Lipid Type | Composition | Types in below |
|---|---|---|
| Fatty acid Other detergents | micelle | Type 1 |
| Diacyl Lipid | bilayer | Type 0 |
| Diacyl Glycerol | inverse micelle | Type 2 |
Core of the membrane is the most hydrophobic environment we can find.
In the core of folded protein, the hydrophobic core coudl have a few polar groups. But not poler group in Membrane core. Which gaves the membrane a barrier property
 |
|---|
| © Chandrashekhar V. Kulkarni |
“Fluid Mosaic” model of biological membranes
 |
|---|
| © Veronika Novotná, et al. |
Living cells actively maintain a certain membrane fluidity by adjusting the composition of the lipids in their membranes.
The lipids are the solvents, the proteins are solutes.
This drawing is wrong. The membrane can pack more protein than it. Exp: Mitochondria.
Secondary Structure Types in Membranes
 |
|---|
| © Guangshun Wang |
| Header One | Header Two |
|---|---|
| Water | |
| Interface | 15 Å |
| Core | 30 Å |
| Interface | 15 Å |
| Water |
Membrane-spanning proteins have very few polar groups groups, such as the peptide back bone exposed to the core of the bilayer.
| Membrane Protein | |
|---|---|
| α-helix |
|
| β-barrel |
|
Membrane integral protein challenges:
- Residues: Hydrophobic residues on the menbrane core area
- Carbon back-bond: Form hydrogen bond with itself.
Typically, the surface of the proteins interacted with membrane are even more hydroponically than the core of folded protein.
30Å membrane core takes about 20 residues of α-helix.
β-barrel only find in gram-negative bacterial, mitochondrial and chloroplasts. (Human, about 50 β-barrel gene and only on mitochondria genome. 2% of gene is β-barrel in E. coli)
Structure of Bovine Rhodopsin
A protin in eye cell membrane.
Salt bridges and hydrogen bonds are rare in membrane protein.
| © Molview, PDBID=1f88 Rhodopsin Dimer |
Structure of Rhodopsin
Spacefilling
- Side View Top View
Structural features of membrane proteins
- Exterior is very hydrophobic over ~30 Å slab of the membrane
- Interior has amino acid composition like that of a soluble protein
- Helices are close-packed
- Salt-bridges and helix-helix hydrogen bonds are rare
- ionizable residues are nearly always salt bridged.
- Helix-helix interactions are mainly driven by van der Waals packing
 |
|---|
| © Yi Xia |
The human erythrocyte protein Glycophorin A has a single membrane-spanning α-helix
Hydropathy Analysis
 |
|---|
| © Yair Benita, et al. |
Take the 19 amino acids in a box from the chain and calculate the hydrophobicity, and them you slid it one by one to make the Sliding Window Hydropathy Plot.
Glut1
| © Molview, PDBID= 4pyp Rhodopsin Dimer |
 |
|---|
| © MobeenRaja, et al. |
Lipids, Membranes and Membrane Proteins|Graduate Biochemistry 7| Tulane
https://karobben.github.io/2021/09/29/LearnNotes/tulane-biochem-7/











