6 Protein Folding and Degradation|Advanced Cell Biology|Tulane
Protein Folding and Degradation
• Lodish MCB, 8th ed., pages 81-105 and 622-631.
• 3.2 Protein Folding
• 3.3 Protein Binding and Enzyme Catalysis
• 3.4 Regulating Protein Function
• 13.6 Transport Into and Out of the Nucleus
Protein Folding
Principles of protein folding:
- AA Seq. → 3D Structure and Function.
- Assisted by: ATP-dependent chaperones and chaperonins
- Misfolded/denatured proteins → Diseasecan
- Exp: amyloid fibril: Alzheimer & Parkinson.
Factors that Affect Protein Folding/Structure
Hypothetical protein folding pathway:
- Primary → Secondary → Tertiary
- Native conformation: lowest free energy
Forces that govern folding
- Hydrophobic interaction (favors)
- Conformational entropy (disfavors)
- Steric hindrance limits dihedral angles off central α-carbon
- van der Waals contacts (packing)
- Hydrogen bonds
- Electrostatic interactions
PS:
- Planar peptide bonds
- φ and ψ: steric clash
PS: How AA Seq determing the high dimensions information
For example, :
- Steric effect: a large side chain, such as that of (W), might sterically block one region of the chain from packing closely against another region
- Charge effect: a side chain with a positive charge (A) , might attract a segment of the polypeptide that has a complementary negatively charged side chain (e.g., D).
- Secondary structure: Example we have already discussed is the effect of the aliphatic side chains in heptad repeats in promoting the association of helices and the consequent formation of coiled coils. Thus a polypeptide’s primary structure determines its secondary, tertiary, and quaternary structures.
Steric hindrance and secondary structure
- Values of dihedral angles φ and ψ are excluded by contact between bulky groups - steric hindrance (clash)
- “Allowed” regions of Ramachandran Plot correspond to major secondary structures
Molecular Chaperone Families
• stabilize unfolded or partly folded proteins and prevent aggregation during synthesis, membrane translocation, assembly, and degradation.
• Molecular chaperones bind exposed segments and surfaces of unfolded proteins.
• Hsp60-Hsp10 family chaperones (Mito and chlp) manage folding of enzymes and proteotoxic stress
• Hsp70-Hsp40 family chaperones (in cyto, ER, nuc,mito, chlp) manage protein translocation across membranes and proteotoxic stress
• Hsp90 family chaperones (cyto, nuc, ER, mito, chlp) regulate signaling and gene expression and manage proteotoxic stress
• Hsp100 (AAA+) family chaperones (cyto, mito, chlp) unfold proteins for degradation and manage proteotoxic stress
• Hsp25 family chaperones (in cyto) manage actin assembly and proteotoxic stress - do not utilize ATP
| Molecular Chaperones | Chaperonins |
|---|---|
| Small protein which bind to a short segment of a target and stabilize unfold or part folded protein. | Large protein which form a folding chamber into which all or part of unfolded protein, give it time and an appropriate environment to fold properly. |
Functions:
- Preventing aggregation
- Fold newly made proteins
- disassmble ptentially toxic protein aggregates duet to …
Chaperones: Hsp60-Hsp10 ATP cycle
 |
|---|
| © Alexandra Richardsona, et al. |
Video:
Hsp60: Huge cylindrical supramolecular assemblies
Two groups:
- Group I: The Prokaryotic Group
- GroEL/GroES
- Group II: The Eukaryotic Group
- TriC
- ATP + GroEL → ADP:
- unfolded protein in
- lid of GroES sealed
- ATP → ADP → protein fold
- ATP → ADP → GroES and peptide release
- Pepetide enter another chamber → Release/Repeats
Chaperonins: Hsp70-Hsp40 ATP-cycle
- Hsp40-bound “client” protein binds to Hsp70
- Hsp40 J-domain triggers ATP hydrolysis, Hsp70 closes, Hsp40 released
- Waiting/folding
- ADP/ATP exchange triggers opening of Hsp70
- Release of client protein
Hsp70
Monomeric
Families of heat-shock protein Hsp70 homologs:
- Hsp70 in Mito; 2. Bip in ER, 3. DnaK in
Heat-shock: large expressed after heat-shock in bacteria.
- +ATP → Open conformation → Expose Hydrophobic Region → Bond client.
- ATP → ADP → Close conformation → helping folding
- ADP Release → conformation change → Target release
- ATP bond → binds another target
- Check:
- appropriate folded → Cannot bind again
- inappropriate folded → bind again
Hsp40
Four main families of nucleotide exchange factors:
- GrpE: bacteria
- BAG; HspBP; Hsp110
Hsp40 (co-chaperone) facilitate Hsp70 processes.
- Increasing the ATP hydrolysis. ×100 ~ ×1000
Hsp90 ATP cycle
- Hs70-bound “client” protein binds to Hsp90
- ATP binding triggers closing of Hsp90
- Waiting/folding
- ADP hydrolysis and release
- Release of client protein Hsp70-bound Hsp70
Dimer protein
Usually recognizing partially folded protein, highly conserved among species.
Four families: two in cytosol, one in Mito, and one in ER.
Important: Helping miss folded protein under the stress, convert them into active form or held in a functional conformation.
- open conformation → Bind target
- +ATP → Closed conformation
- ATP → ADP → target release
Else:
- Cooperate with Hsp70
- Influenced by covalent modification
- Facilitate identification and degradation
AAA+ Protein Family
Aggregate disassembly
- Hsp100 of yeast
- ClpB of E. coli
Vesicular transport complex disassembly
- NEM-sensitive factor for SNARE disassembly
- Vps4 for ESCRT disassembly
DNA-binding protein assembly
- Clamp loader
Export of mRNA from nucleus- Dbp5
Protein translocation across membranes- p97 for ER-associated degradation (ERAD)
Protein degradation- ClpA, ClpX of E. coli
- “Cap” of 26S proteasome (6 in 19S cap)
Molecular motor- Dynein for organellar transport along microtubules
(ATPases associated with various cellular activities, homologous to hexameric nucleic-acid helicases)
3.4 Regulating Protein Function
- At the level of synthesis, degradation, or through noncovalent or covalent interactions.
- Proteins marked for destruction with a polyubiquitin tag by ubiquitin ligases are degraded in proteasomes.
- Allosteric mechanisms act as switches, reversibly turning protein activity on and off.
- Higher-order regulation includes the intracellular compartmentation of proteins.
- Proline cis/trans isomerizations.
- Alters structure of a protein SH2 domain.
- Can influence protein activity.???
- Proline isomerase may act as switch to regulate protein activity.
From Textbook:
- ALtering the rate of protein synthesis and degradation
- Change the intrinsic activity, Exp: active or in active conformation
- Change the location and the concentration within the cell.
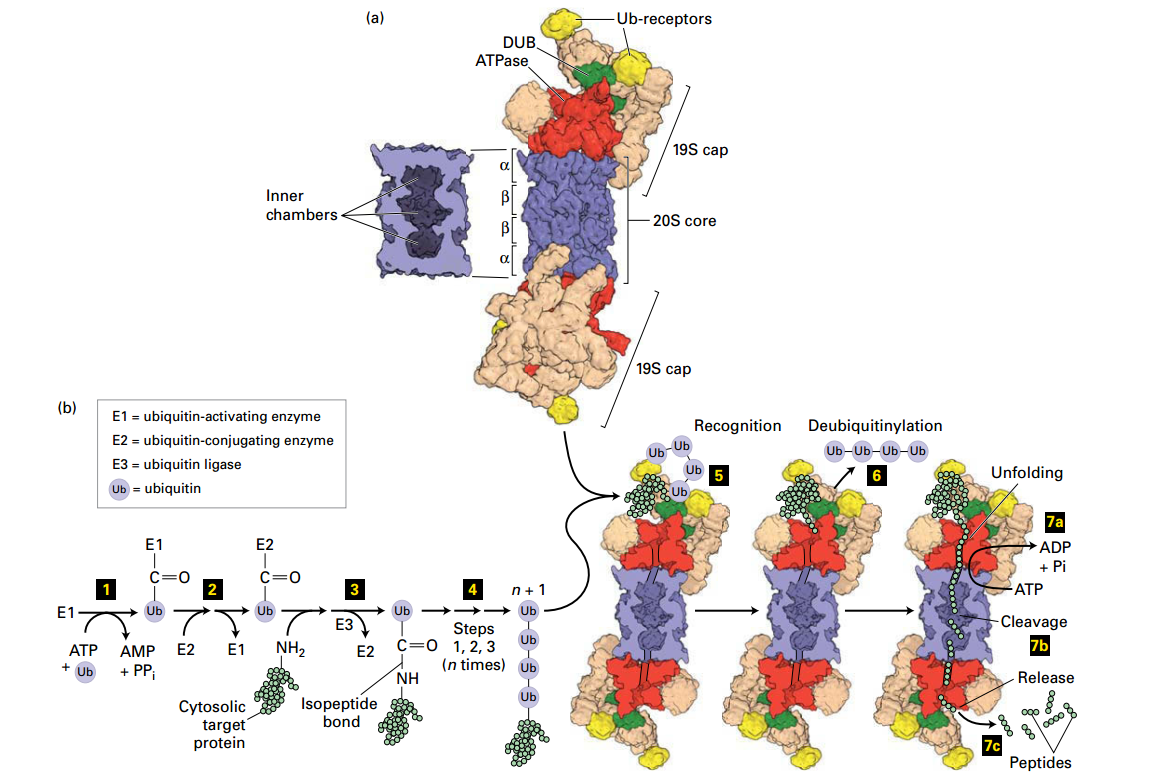
Ubiquitin- and Proteasome-Mediated Proteolysis
 |
|---|
| © Lodish, Molecular Cell Biology, Eight Edition, p98 |
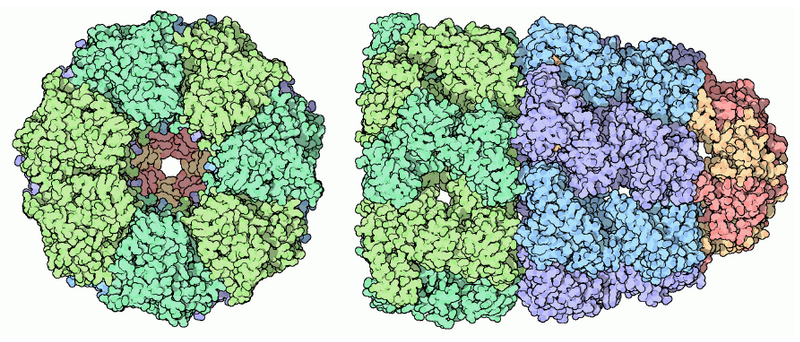
Proteasome
Structures:
- ATPas: AAA+ family: Red
- Ubiquitin recepor: Yellow
- Deubiquitinase Enzyme: Green
Ubiquitin and Formation
| Ubiquitin’s type | Features |
|---|---|
| Monoubiquitinylation | Single Ubiquitin |
| Multyubiquitinylation | Multiple, single ubiquitin |
| Polyubiquitinylation | A polymeric chain of ubiquitin |
Degradation in Proteasome
- Actication: ATP + ubiquitin (Ub) → activate Enzyme E1
- E1 - Ub
- Ttransfers Ub → E2 (step 2 )
- E2 - Ub
- Formation: Ubiquitin ligase (E3) transfer Ub → Lysine–NH2 and forming an isopeptide bond
- UB - Peptide
-
- more Ub by repeating steps 1 – 3
- n * (Ub) - Pepide
- polyubiquitinylatd targed → Ub receptors (19S cap)
- went into proteasome
- deubiquitinase enzyme (19S cap) → Ub remove
- a. +ATP → unfold → the 20S core (
b. coordinately with step 6: protein → short peptide → release (7c)
Degradation: Proteosome recognition, 1. de-Ub protein, 2. unfolds protein; protein transferred to core proteolysis chamber, protein cleaved into short peptides (2-24 aa) and released for further degradation by soluble proteases.
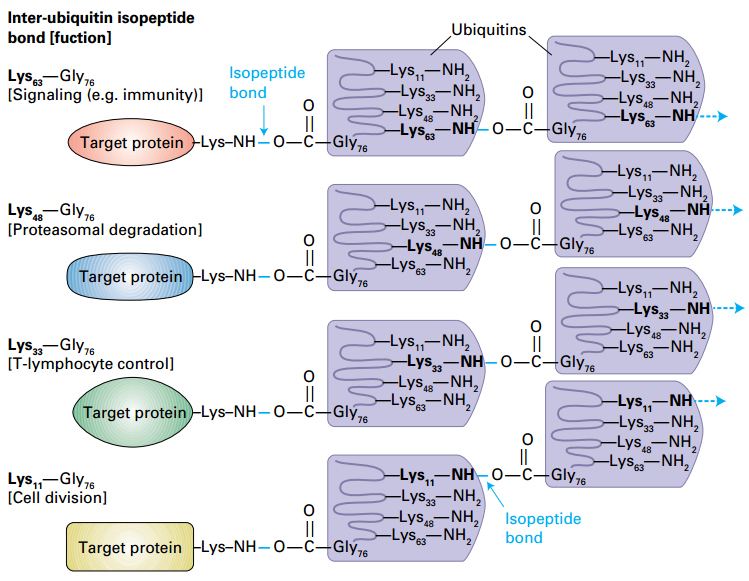
Determination of polyubiquitin function by the lysine used for inter-ubiquitin isopeptide bonds
 |
|---|
| © Lodish, Molecular Cell Biology, Eight Edition, 104 |
Signaling: Lys-63;
Proteasomes: Lys-48:Gly-76 isopeptide.
T-lymphocyte control: Lys-33
Cell division: Lys-11
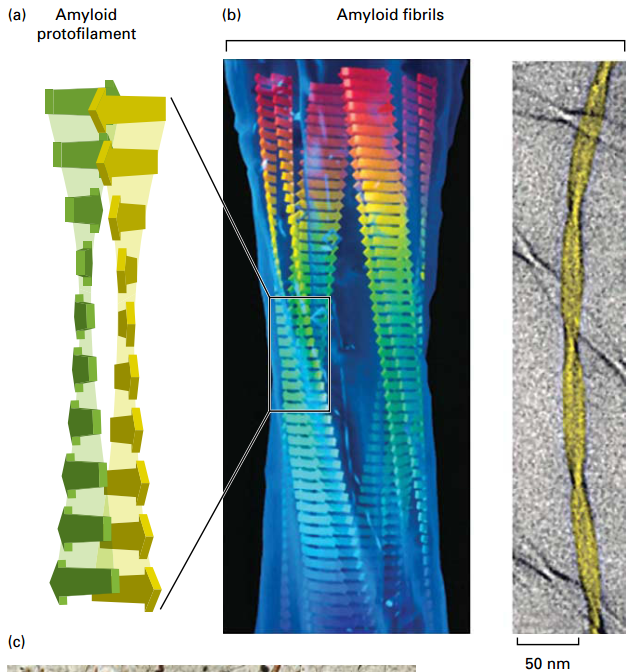
Misfolded Proteins can form Amyloid Aggregates
 |
|---|
| © Lodish, Molecular Cell Biology, Eight Edition, p88 |
- Amyloid aggregate plaques form inside or outside many cell types.
- Flat sheets form protofilaments, assemble into thicker fibrils, and into larger aggregates or macroscopic plaques.
- Cause neurodegenerative amyloidosis diseases, including Alzheimer’s disease, Parkinson’s disease, and transmissible spongiform encephalopathy (“mad cow” disease).
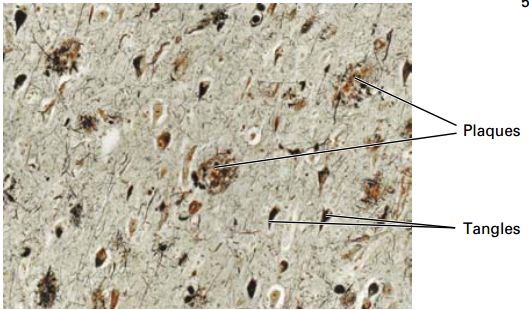
Clinical Case: Alzheimer’s Disease
 |
|---|
| © Lodish, Molecular Cell Biology, Eight Edition, p88 |
68 yo male seen in ER with apparent confusion, disorientation of where he is or the present date.
• Most common form of dementia.
• Affects 1 in 10 people over 65 and 50% of people over 85.
• Plaques (Aβ peptide) and tangles (tau microtubule-associated protein) results in gradual and progressive loss of neurons and cognitive function over a period of 4-15 years (memory lapse, language impairment, loss of motor functions).
Alzheimer’s Disease Fact Sheet; 2021 NCBI
Prion Diseases & Transmissible Spongiform Encephalopathies
TSE - spongiform neurodegeneration, astrocytic gliosis.
Prion - proteinaceous infectious agent, considered heretical concept for transmission.
Lehninger Box 4-6
Transmissible Spongiform Encephalopathies
- Creutzfeldt-Jakob Disease (CJD)
- Gerstmann-Straussler-Scheinker syndrome (GSS)
- Fatal Familial Insomnia (FFI)
- Kuru: Fore tribe in New Guinea
- Wild Animals and Livestock (bovine, mink, feline, elk, sheep)
The Prion Hypothesis for TSEs
PrPSc + PrPc → 2PrPSc
| Synfrome | Etiology | |
|---|---|---|
| Sporadic | sCJD | Spontaneous conversion of PrPc to PrPsc (usually not mutant) |
| Acquired | Kuru | Ritualistic endocannabilism |
| iCJD | Exposure to contaminated growth hormone, gonadotrophins, dura mater, cornea or surgical instruments | |
| vCJD | Ingestion of BSE-contaminated food products | |
| Familial | fCJD | Germline mutation in PRNP locus |
| GSS; FFI |
From: Hilton (2006) J Pathol 208: 134, Table 1 Iatrogenic CJD 2004
Diseases involving Amyloid Formation: Know the normal and abnormal proteins, the disease, and specific mechanisms.
| Disease | Type II | Alzheimer’s disease | Prion disease | Huntington’s disease | Parkinson’s disease | ALS | Spinal Cerebellar Ataxia |
|---|---|---|---|---|---|---|---|
| Protein | Amylin | Amyloid Precursor Protein (APP) | PrPc | Huntingtin protein | α-synuclein | SOD | ataxin |
| Function | Glycemic regulation with insulin | ECM, migration during development | Unk. | Unclear, MAP, vesicle trafficking | Unclear, MAP, vesicle trafficking | Destroy oxygen radicals | Unclear |
| Abnormal protein | n/a | Aβ42(β-Amyloid) | PrPsc | mHTT, Trinucleotide repeats (polyQ) | Lewy bodies | n/a | Trinucleotide repeats (polyQ), and nonPolyQ |
| Effects | Apoptosis of β islet cells | Neuronal death | Neuronal death | Neuro-tramsmitter release, neuronal death | Neuro-transmitter release, dopaminergic neuronal death | Dementia neuronal atrophy | Neuronal deat |
13.6 Transport Into and Out of the Nucleus
 |
 |
|---|---|
| © Jeroen Vangindertael, et al; 2018 | © Roderick Lim, et al; 2006 |
 |
 |
| © Mario Tagliazucchi, et al; 2015 | © Anita H Corbett, et al; 2004 |
• Unidirectional transport of a protein larger than 40 kDa through large, complex nuclear pore complexes requires a nuclear-localization or nuclear-export signal, nuclear transport receptors, Ran G-proteins, and localized Ran?GEFs and GAPs.
• Other molecules, including mRNPs, are transported by a Ran-independent pathway.
Nucleoporins:
- Structural nucleoporins (circule the memrain)
- Membrane nucleoporins (Y complex)
- FG nucleoporins (matrix on the pour center)
Transport Into and Out of the Nucleus
-
The Nu Env: NPCs, ×30 different nucleoporins.
- FG-nucleoporins: multiple repeats of a short hydrophobic sequence (FG-repeats), line the central transporter channel and play a role in the transporter.
-
>40 kDa: nuclear transport receptors interact
-
nuclear-localization signal (NLS) or a nuclear-export signal (NES) on the proteins in/out.
- NLS: Nucleus-restricted proteins
- NLS+NES: proteins that shuttle between the nucleus and cytoplasm
-
Each type of NESs/NLSs is thought to interact with a specific nuclear transport receptor.
-
Transient interactions: Protein-[NES/NLS] + receptor.
- Transport receptors + FG-repeats: very rapid diffusion through the NPC,
-
Ran: a monomeric G protein
- Localization of the Ran guanine (Ran-GEF) in the nucleus and of the Ran GTPase-activating protein (Ran-GAP) in the cytoplasm creates a gradient with high concentrations of Ran⋅GTP in the nucleoplasm and of Ran⋅GDP in the cytoplasm.
- The interaction of a cargo complex with Ran⋅GTP in the nucleoplasm causes dissociation of the complex, releasing the cargo into the nucleoplasm, whereas the assembly of an export cargo complex is stimulated by interaction with Ran⋅GTP in the nucleoplasm.
-
Most mRNPs are exported from the nucleus by binding to a heterodimeric mRNP exporter. RNA helicase associated with the cytoplasmic filaments of the NPC that removes the heterodimeric mRNP exporter once the transport complex has reached the cytoplasm.
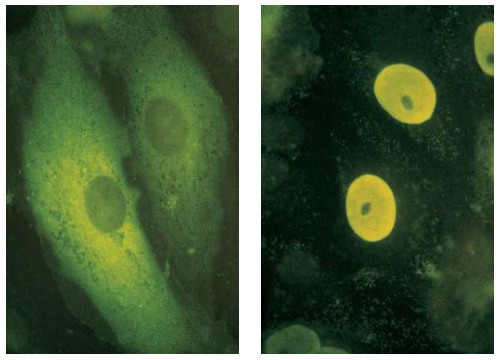
Nuclear-Localization Signals (NLSs) Direct Proteins to Cell Nucleus
 |
|---|
| © Kalderon D, et al; 1984 |
NLSs is a 7 residue sequence rich in the terminal of the protein, exp: PLLLRLV
- All nuclear proteins come from cytosol.
- They all have NLSs
- Experiment:
- (a) Pyruvate kinase: in the cytosol for glycolysis.
- (b) Chimeric pyruvate kinase containing SV40 NLS at its N-terminus: transported into the nucleus.
Nuclear Import
 |
|---|
| © Terence David Allen, et al. 2000 |
Ran & nuclear transporter receptor was purified.
In Cytosol:
- Importin + NLS-[Cargo].
- Importin-NLS-Cargo passively diffuses.
In nucleus:
- Ran-GEF: Ran-GDP + GTP → Ran-GTP + GDP
- Importin binds Ran-GTP → Cargo release.
- Importin-Ran-GTP diffuses through NPC.
Back in cytosol.
- Ran-GTP reacts with Ran-GAP → part of cytoplasmic filament.
- GTP hydrolysis → low affinity for Importin → releases Importin.
RanL a small monomeric G protein: GTP/GDP-bound conformation: supply the energy
NTR binds to the NLSs: Recognition
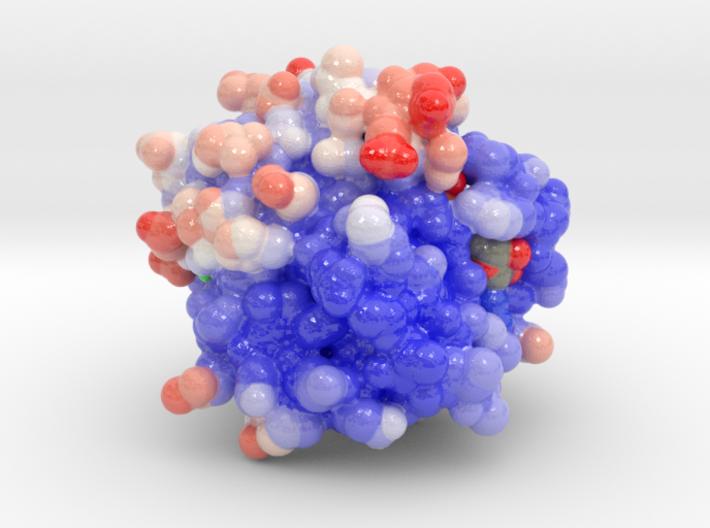
Monomeric G Proteins: The GTPase Switch
 |
|---|
| © biologicmodels |
- GTPase:
- GTP-bound: Active “on” conformation → interacts with target proteins to regulate their activities.
- GDP-bound: inactive “off” conformation → intrinsic GTPase activity → hydrolyzes GTP to GDP.
GEF: Guanine nucleotide exchange factor, stimulates exchange of GDP-(off) to GTP-bound (on) forms.
GAP: GTPase-activating protein, stimulates GTP (on) hydrolysis to GDP (off).
Examples: Ras, Ran, Rho, ARF
Ran-Dependent Export
 |
|---|
| © Terence David Allen, et al. 2000 |
In Nucleus:
- [Ran-GTP]-[Exportin]-[Cargo] → passive diffusion.
In Cytosol:
- Ran-GAP: [Ran-GTP]-[Exportin]-[Cargo] → Ran-GTP + Cargo + Exportin
- Passively diffuse: Ran-GDP & Exportin → into Nucleus.
Back in nucleus:
4. Ran-GEF: Ran-GDP + GTP → Ran-GTP + GDP
Ran-Independent mRNA Nuclear Export
NXF1: Nuclear Export Factor 1
NXT1: Nuclear Export Transport 1
mRNP: Messenger Ribonuclear Protein Complex
Nucleoplasm:
- Passive diffusion: Heterodimeric NXF1/NXT1 + mRNPs.
Cytosol - Dissociation: RNA helicase (Dbp5) + ATP + [NXF1/NXT1]-[mRNP] → NXF1 + NXT1 + mRNA + ADP + Pi
- Recycling: Ran-dependent import recycles NXF1 and NXT1
Vesicular Traffic
Class I Presentation to CO8⁺ T Cells [go to find the pic and make more notes.]
Cross presentaiton
P: 1110
Class II Presentation to CD4⁺ T Cells
P: 1112
Lodish MCB, 8th ed., pages 631-659.
Techniques for Studying the Secretory Pathway
Protein Transport through the Secretory Pathway
Secretory and Endocytic Pathways
Sec Mutants Identify 5 Stages of Secretory Pathway
Exam: Classes , what machine could be broken/mutant
Coated Vesicles Involved in Protein Trafficking
Coated Vesicles table, 1, 2, 4
Docking/Fusion of Vesicle w/Target Membrane
Coil-Coil structure is really staby, not easty to recycle.
KDEL Receptor Retrieval of ER-Resident Luminal Proteins from the Golgi
slide difference pH between in cis and trans Golig: help the cargo binds and releaes (pH trigger).
Dynamin Pinching Off of Clathrin-Coated Vesicles
High energy state to close the bud neck
14.2 Molecular Mechanisms of Vesicle Budding and Fusion
14.3 Early Stages of the Secretory Pathway
14.4 Later Stages of the Secretory Pathway
 |
|---|
| © Keith P. Delaune; Khalid Alsayouri. |
Exam:
Draw cell component
G protein regulation and fedality
6 Protein Folding and Degradation|Advanced Cell Biology|Tulane
https://karobben.github.io/2021/09/30/LearnNotes/tulane-cellbio-6/