Protein Translocation
• Lodish MCB, 8th ed., pages 583-622. • 13.1 Targeting Proteins To and Across the ER Membrane
• 13.2 Insertion of Membrane Proteins into the ER
• 13.3 Protein Modifications, Folding, and Quality Control in the ER
• 13.4 Targeting of Proteins to Mitochondria and Chloroplasts
• 13.5 Targeting of Peroxisomal Proteins
Protein translocation was also called protein sorting/protein targeting.
Two types of protein sorting:
- single-based targeting
- Newly or completed transcriptions
- Leads insertion in the membrane, trans-membrane into organelles
- vesicle based trafficking.
- Secrete protein, Glogi, lysosome, and plasma membrane
Targeting Proteins To and Across the ER Membrane
All soluble protein are initially delevered into ER.
Two main types of protein: Soluble protein and Integral protein.
Synthesis of:
-
secreted proteins
-
integral plasma-membrane proteins
-
proteins destined for the ER, Golgi complex, plasma membrane, or lysosome begins on cytosolic ribosomes.
-
A signal sequence, SRP, and SRP receptor system docks the ribosome on an ER translocation and cotranslationally inserts the nascent protein into or through the ER membrane.
Structure of the Rough ER
- Convoluted membranes continuous with nuclear membrane.
- sER: Cellular lipid synthesis (sER).
- rER: First step in secretory pathway – rough ER – with attached ribosomes.
- Synthesis and processing for plasma membrane and secreted proteins, and proteins functioning in the ER, Golgi complex and lysosomes.
- Protein synthesis on the ER:
- ER membrane-bound and free cytosolic ribosomes are identical.
- Membrane-bound ribosomes: recruited to the ER during synthesis of a polypeptide containing an ER signal sequence.
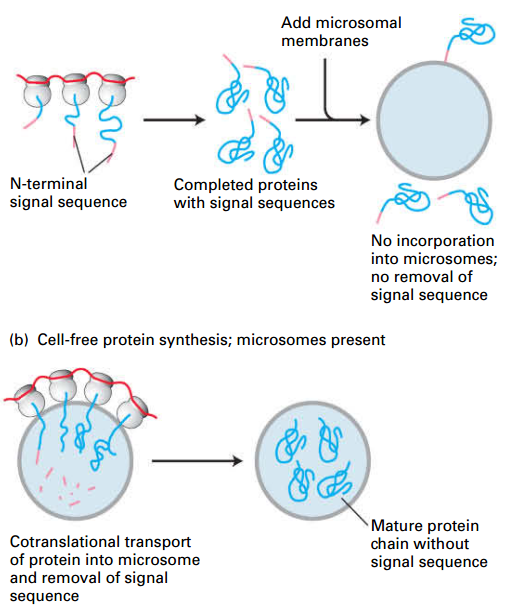
Secretory Proteins Enter the ER Lumen
- Pulse-chase experiments: secretory proteins are in ER lumen right after synthesis.
- before the synthesis of first 70 AAs.
- Pulse radiolabeled amino acids
- only newly synthesized proteins labeled.
- Cells homogenized: shears rough ER into small vesicles called microsomes.
- Purified microsomes treated with a protease: (–) Detergent: proteins in ER are protected from digestion. (+) Detergent: dissolves ER membrane; proteins not protected from digestion.
- Detergent: disrupt the membrane system to expose the secrete protein
- Conclusion: newly made proteins are inside the microsomes (lumen of the rough ER) after synthesis.
Simultaneous Translation / Translocation
- Hydrophobic N-terminal signal sequence targets secretory proteins to ER membrane during translation from free cytosolic ribosomes.
- Initiates translocation of the growing polypeptide across the ER membrane
- Signal sequences cleaved from most secretory proteins after translocation.
- Cell-free experiments demonstrate that translocation of secretory proteins into microsomes is coupled to translation – cotranslational insertion
Cotranslational Translocation
- SS transcription: N-terminal ER signal sequence (SS) emerges from the ribosome first.
- SRP recognition: Signal recognition particle (SRP) binds SS – arrests protein synthesis.
- SS, SPR, Ribosome; SRPr complex: SPR-ribosome complex binds SRP receptor in ER membrane; interaction strengthened by the binding of GTP to both the SRP and its receptor.
- SRP dissociates: Transfer of nascent polypeptide–ribosome to translocon; opens translocation channel. SRP and SRP receptor both hydrolyze bound GTP; dissociates SRP from ribosome and receptor; restarts protein synthesis.
- SS cleaved: Peptide passes thru translocon into ER lumen; signal sequence cleaved.
- extrudes: Growing peptide chain extrudes thru translocon into ER.
- Ribosome Dissociated: Translation finishes; ribosome is released
- Folded: Translocated protein folds into native conformation and translocon closes.
Nucleoside hydrolysis reactions that regulate processes using a timer-like function
| GTP |
ATP |
| Ef-tu in protein synthesis G-protein signaling Ran in nuclear import/export SRP and SRP receptor Tubulin dynamic instability Sar1 in vesicle budding Rabs in vesicle fusion Dynamin in bud neck closure |
Actin for assembly Hsp70 for client binding/release Glucokinase for positive cooperativity |
Post-translational Translocation
BiP: (Hsp70 family ATP-dependent molecular chaperone) BiP bind peptide could prevent it’s backward sliding.
- Direct interaction: SS + translocon. N-terminal segment of the protein enters the ER lumen; signal peptidase cleaves SS.
- BiP + Sec63 complex: provide driving force for unidirectional translocation across the ER membrane.
- Random inward sliding: exposes more nascent protein.
- BiP-ADP binding: ratchets nascent protein into ER. BiP ATP hydrolysis essentially powers protein translocation across membrane.
- Releases: ATP for ADP exchange releases BiP from nascent protein.
- Folding: Protein folds into native conformation.
13.2 Insertion of Membrane Proteins into the ER
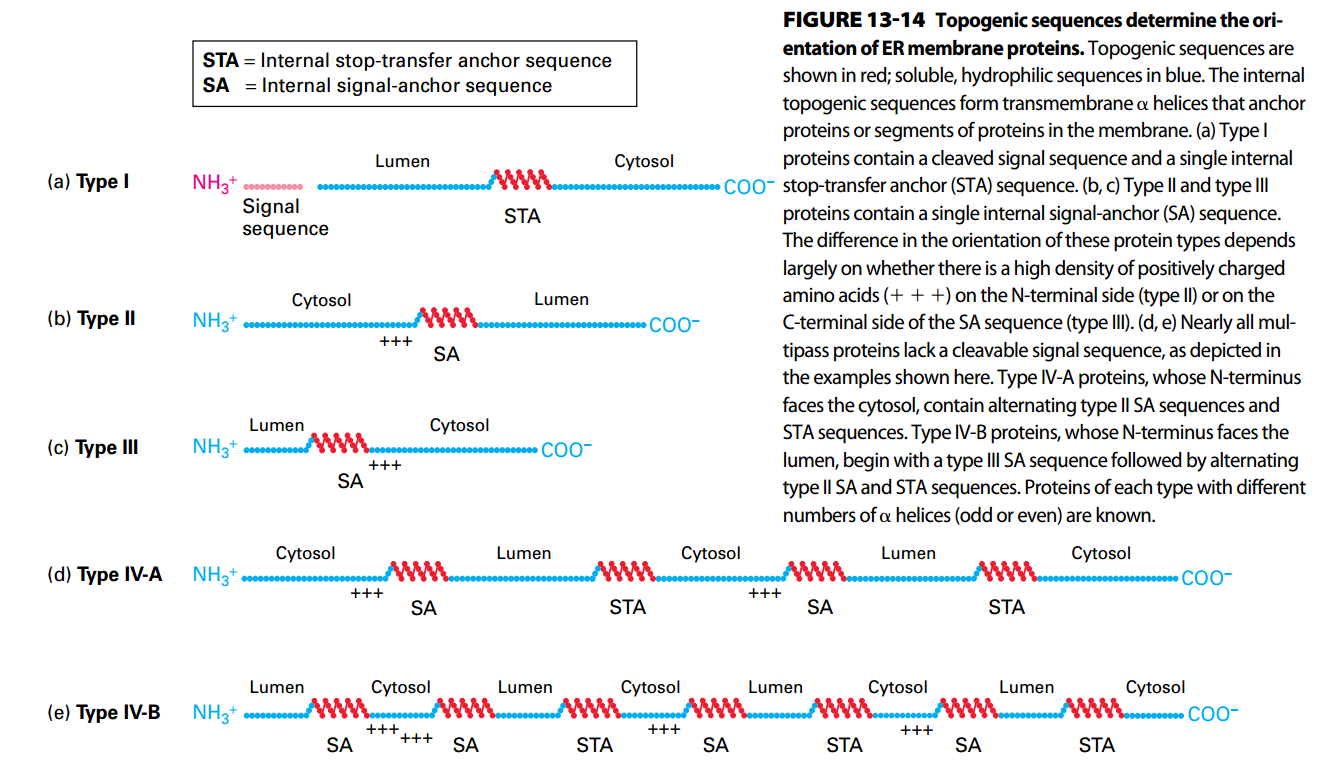
- Topogenic sequences:
- N-terminal signal sequences
- Internal sequence
- internal stop-transfer anchor sequences
- internal signal-anchor sequences
- direct the insertion of nascent proteins into the ER membrane.
- Topogenic sequences can be predicted by computer by identify hydrophobic topogenic segments.
- GPI anchor: Initiate as Type I but cleaved and transferred into GPI molecue
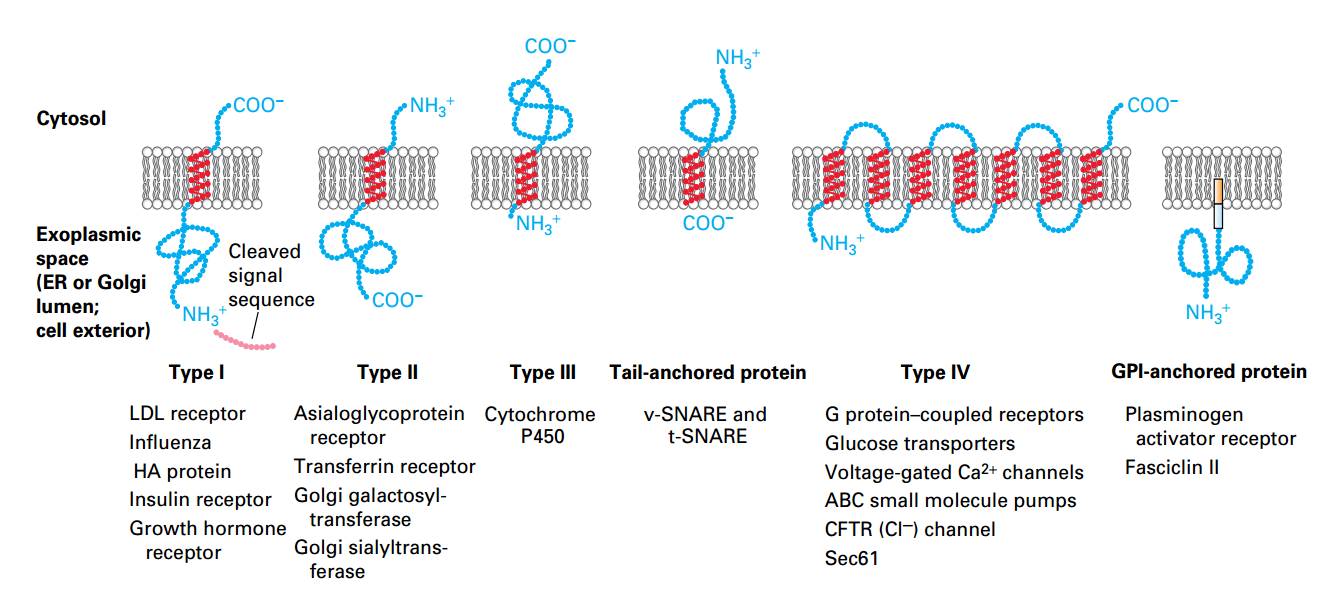
Classes of ER Membrane Proteins
- Classified by
- Orientation
- Locations of N/C-termini
- the Types of targeting signals.
- α helices segments (20- 25 A, Hydrophobic) → Embed.
- Hydrophilic regions fold into conformations outside membrane.
- Type IV: n × α helices.
- GPI anchor: protein in membrane by phospholipid anchor.
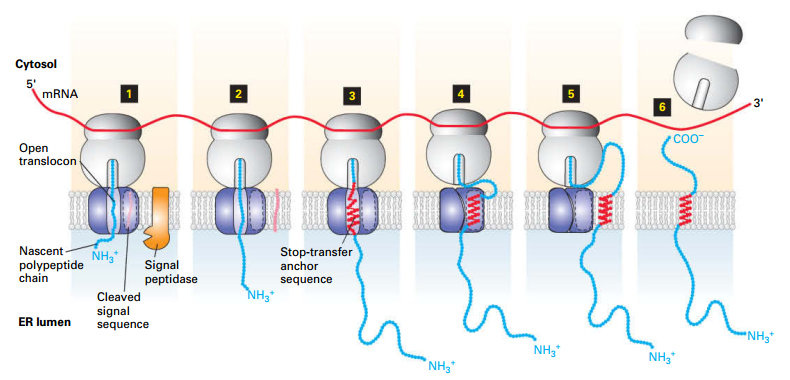
Type I Insertion/Orientation
Topology:
| ER Lumen |
Transmembrane Domain |
Cytosol |
| N- |
Single |
-C |
- Translocation initiation and Ss cleavage.
- Ppeptide elongates.
- Elongation until hydrophobic stop-transfer anchor sequence enters translocon → prevents further translocation.
-
- Stop-transfer anchor sequence (α-helix) moves laterally into membrane.
- Translocon closes.
- Newly synthesized AA remain in cytosol.
- Synthesis completes; ribosomal subunits are released.
PS: cotraslation translocation
Sec61α domain, plug peptide.
Exp: HGH receptor
Types II and III Insertion/Orientation
- Have single internal hydrophobic signal-anchor sequence (SAS; serves as both ER signal sequence and membrane anchor).
| Category |
Type I |
Type II |
| N-terminal Ss |
None |
None |
| SAS |
Yes |
Yes |
| Positive charges adjacent to SAS |
towards cytosol |
towards cytosol |
| SRP binds |
internal SAS |
internal SAS |
| Positively charged residues |
N |
C |
| cytosol |
C |
N |
SAS functions both Ss & SAS, which is recognition and anchor.
Type II: Start as Type I but not be cleaved, as a result, the N terminal still on the cytosol side.
Type III: N-terminal was anchored immediately since the SAS is so closes to N-terminal. As a result, rest of sequence can’t enter the lumen.
Topology/Orientation of ER Membrane
Red: Topogenic Sequences
Blue: Soluble, hydrophilic sequences
The high dense charged residues adjacent to SA were in the cytosol side.
GPI-Anchored Proteins
|
|
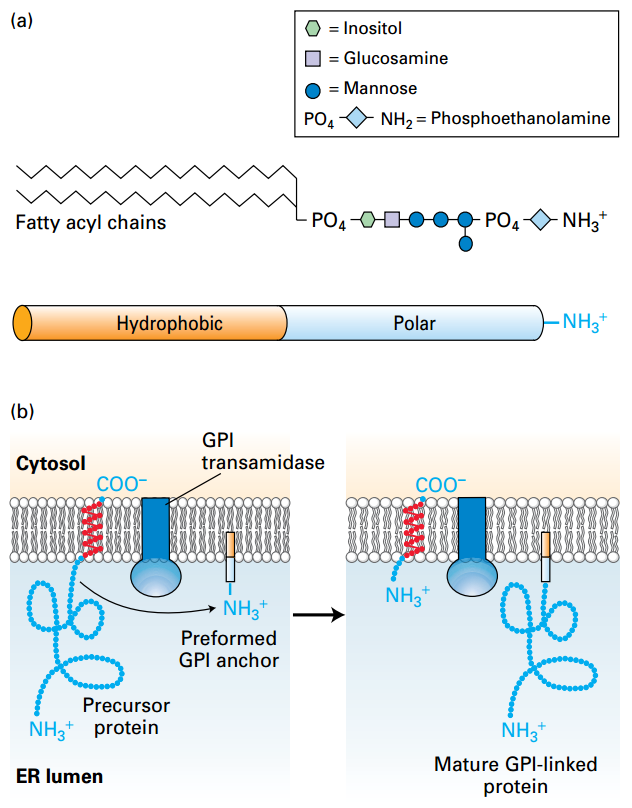 © Lodish, Molecular Cell Biology, Eight Edition, p599 © Lodish, Molecular Cell Biology, Eight Edition, p599 |
GPI anchor covalently attaches protein in extracellular plasma membrane. (a) GPI molecule: hydrophobic fatty acyl chains with polar phosphate groups and carbohydrate moieties. (b) GPI-anchor: 1. Type I protein was synthesized. Transmembrane domain was cleaved and transferred into GPI by Transamidase. Biological Functions: 1, Higher mobility; 2, Not cytosol side residues which might anchor to cytoskeleton; |
GPI: Amphipathic glycosylphosphatidylinositol; Amphipathic
GPI Transamidase: remove the membrane span to the GPI-linked protein.
Tag contacts with ER would transfer to the cell surface through body infusion.
13.3 Protein Modifications, Folding, and Quality Control in the ER
- Oligosaccharides, disulfide bonds, and prolyl isoform modification; Cleavage
- ER chaperones + Lectins:
- help & accelerates correct folding,
- transport to the cytosol to ubiquitin.
- Hinder: Only good protein → Golgi complex in vesicles.
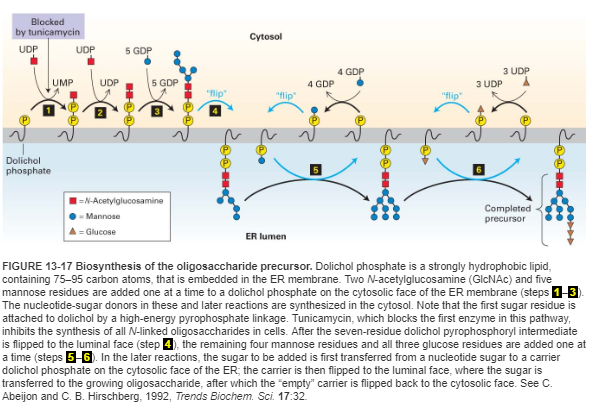
N-Linked Oligosaccharide Precursor Biosynthesis
- N-linked oligosaccharide → RER.
- Oligosaccharide precursor + dolichol phosphate.
- Sugar residues added sequentially as activated sugar-NDPs.
- Steps 1-3: Two N-acetylglucosamine (GlcNAc) and five mannose residues added on ER membrane cytosolic face.
- Step 4: intermediate is flipped to ER membrane luminal face.
- Steps 5-6: Additional sugar-NDPs flipped to the luminal face and transferred to the growing oligosaccharide precursor.
- Tunicamycin blocks step 1, inhibits all N-linked glycosylation.
PS:
- Precursor contain 14 residues and very conservative among eukaryotic.
- Function: Glc₃Man₉(GlcNAc)₂
- formation: Dolichol Phosphate -[pyrophosphate bond]- GlcNAc₂ (Core)→ Man₉ (Branches) → Glc₃ (Tag)
- Asn-X-Ser/Thr: Not all, Not predictable.
- Glycosidase: remove Glc₃ → Act as a signal that the oligosaccharide is complete and ready to be transferred to a protein.
Mucose; extra cellular protein have abundant of glycosylated residues.
Glucose is important: ER quality contra mechanisms for protein folding.
- Protein is maturing, Glucose are being kicked out.
- Protein has be sensed weather it is folded completely.
- The fate was determined by the glucose.
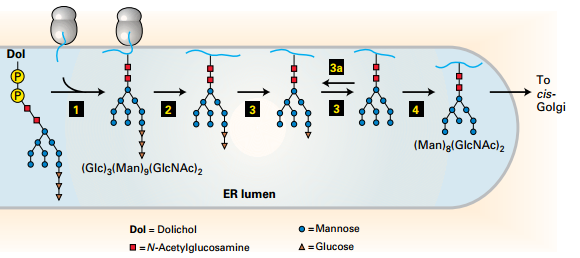
N-Linked Glycosylation and Processing
- Promotes protein folding, stability, adhesion, and recognition.
- Step 1: Oligo precursor transferred into N residues(Asn-X- Ser/Thr)
- Steps 2–4: Remove: Glucose and mannose residues (plays a role in the correct folding of many ER proteins).
- Transfer the protein from ER to Golgi.
- Misfolded proteins delocated into lumen into the cytosol for degradation.
PS:
Examples
- Leukocytes → Cell adhesion molecules
- ABO blood system
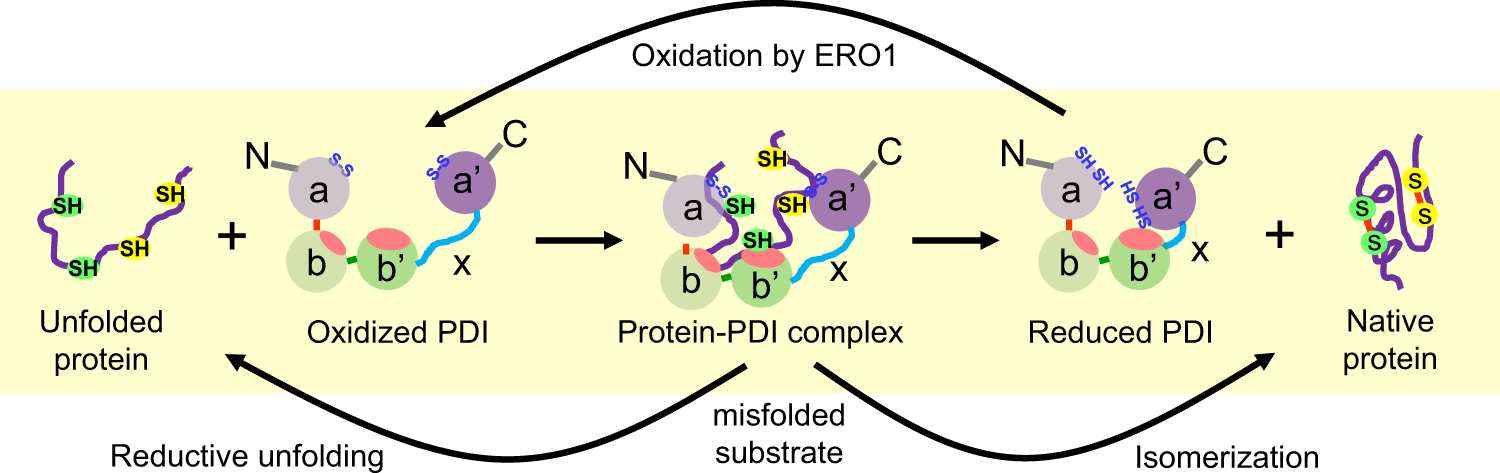
Protein Disulfide Isomerase (PDI)
- Disulfide bonds rearrangement.
- Active site contains two cysteines that can have reduced dithiol or oxidized disulfide forms.
- Functions:
- Formation: Forms disulfide with PDI. Substrate second thiol forms disulfide within substrate, releasing reduced PDI. Reduced PDI is oxidized by Ero1, reforming active site disulfide bond.
- Rearrangement: Reduced PDI reacts similarly to rearrange disulfide bond within substrate. Reaction is repeated until substrate is folded into thermodynamically most stable conformation.
thiol group: sulfhydryl group.
Lectins: carbohydrate binding protein calnexin (On the membrane) and calreticulin (in the lumen) bind selectively to certain N-linked oligosaccharides to help protein folding, reducing peptides aggregate.
Only in the ER:
- for a disulfide bond, Is a mixed disulfide bond, using disulfide bond to connect.
- Correct disulfide bond in cell.
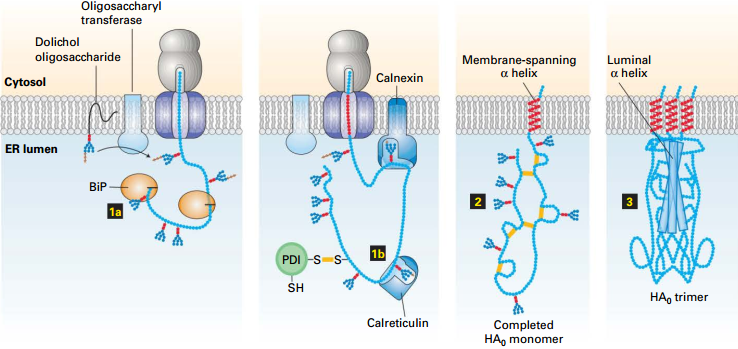
Summary: Hemagglutinin (HA) Folding/Assembly
|
|
 © Lodish, Molecular Cell Biology, Eight Edition, p605 © Lodish, Molecular Cell Biology, Eight Edition, p605 |
1. BiP, and Calreticulin bind nascent protein to prevent premature folding into incorrect conformation. 2. PDI → disulfide bonds → Folding allows trimer formation 3. Complete influenza virion with HA₀ (HA₁ + HA₂) trimer spikes protruding from viral membrane surface. |
Peptidyl-Prolyl Isomerase (PPI) of X-pro peptide bonds
- Accelerates rotation about peptidyl-prolyl bonds in unfolded segments of a polypeptide
- Rate-limiting step in the folding of protein domains
Ire1: The Unfolded Protein Response
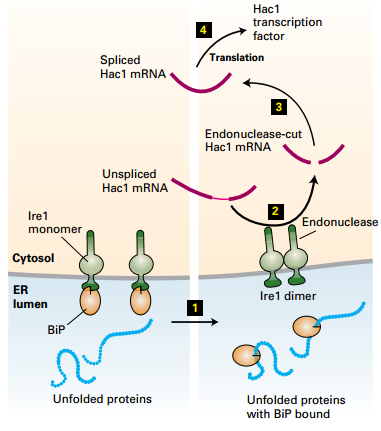 |
1. [Bip]-[Ire1]: heterodimer: Ire1 homodimer was prevented 2. Unfolded protein↑ → Bip↓ : [Bip]-[Ire1] → Bip + Ire1 3. Ire1 + Ire1 → [Ire1]₂ homodimer → Unspliced Hac1 mRNA → Spliced Hac1 mRNA → Translation → Hac1 TF → Nuclear → Gene expression |
| © Lodish, Molecular Cell Biology, Eight Edition, p606 |
|
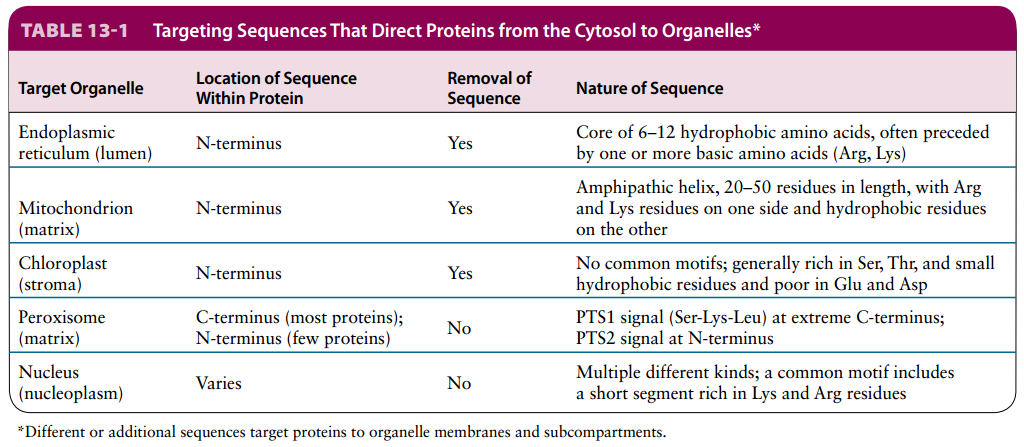
13.4 Targeting of Proteins to Mitochondria and Chloroplasts
- States: Mitochondrial and chloroplast proteins encoded by nuclear genes are synthesized on cytosolic ribosomes, and maintained in an unfolded state by chaperones.
- Targeting Sequences: N-terminal targeting sequences direct post-translational transport of the unfolded proteins through translocons into the organelles.
- Direction: Multiple internal targeting sequences direct protein targeting inside the organelles to different membrane and lumen destinations.
- PS: Energy required, usually two system: one for targeting the organelles, one for sorting to the part of the organelle.
Organelle targeting sequences are similar in their location and general function, but differ in sequence characteristics.
Import of Mitochondrial Proteins
- N-terminal targeting sequence
- Variety: share common motif but different in sequence
- Composition: rich in hydrophobic (A, L), + charged, and hydroxlated (S, T). But lack of - charged (D, E).
- Property: Amphipathic α-helix → one side hydrophobic, one side opsitive charged.
- Required for uptake:
- Matrix-targeting sequence
- Respiring mitochondria (proton-motive force)
- ATP
- cytosolic chaperone proteins that maintain the precursor proteins in an unfolded conformation.
Import into Mitochondrial Matrix
|
|
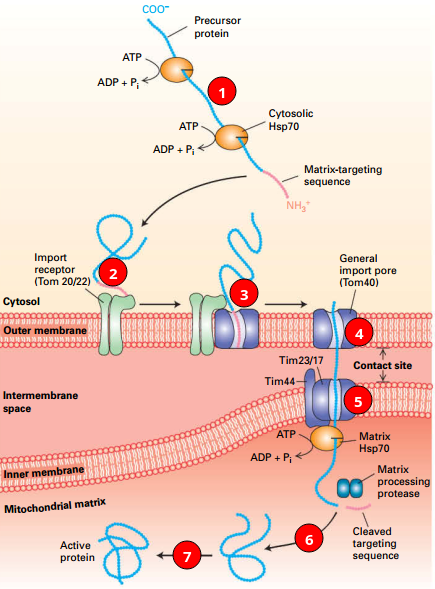 © Lodish, Molecular Cell Biology, Eight Edition, p611 © Lodish, Molecular Cell Biology, Eight Edition, p611 |
1. Cytosol: Protein + chaperones. (Hsp70, Hsp90) 2. Initiating: MTS + import receptor. 3. MTS → outer membrane translocon (Tom40, general import pore). 4. Protein → Tom → inner translocon (Tim). 5. Matrix Hsp70+Cargo: Import↑. 6. [Hsp70]-[Cargo] + ATP → ADP + Pi + Cargo; Targeting Sequence Cleaved 7. Folds (may use other chaperones). |
Matrix-targeting sequence (MTS)
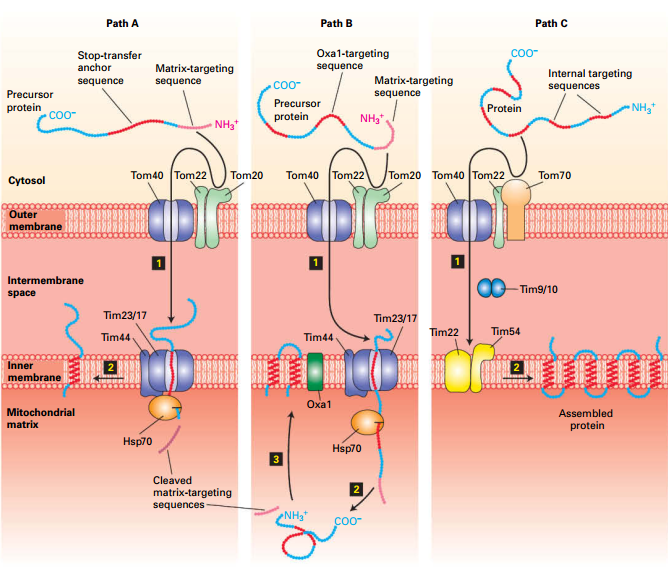
Three Pathways to Inner Mitochondrial Membrane
| Path |
Signal |
Receptor |
Insertion |
| Path A |
N’ MTS |
Tom and Tim |
Stop-transfer anchor sequence: Tim block Transfer |
| Path B |
N’ MTS |
Tom and Tim |
Oxa1: insert protein |
| Path C |
Internal Sequence |
Tom |
Different Tim incorporates multiple hydrophobic segments |
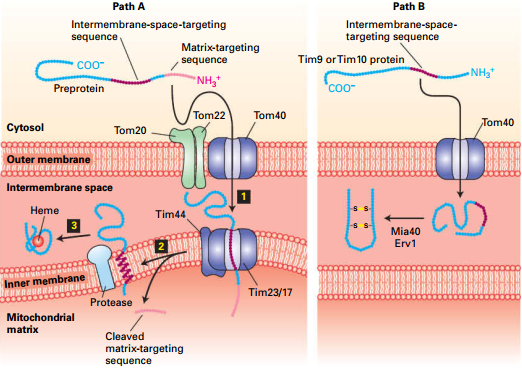
Two Pathways to Mitochondria Intermembrane Space
|
Path A |
Path B |
| Signal Sequence |
MTS (removed in matrix) |
No MTS or Tim targeting sequences. |
| Inner membrane |
Stop transfer sequence → releases from Tim |
Don’t involved |
| Release |
Inner membrane protease |
Stay at here and form disulfide bonds to prevent reverse translocation |
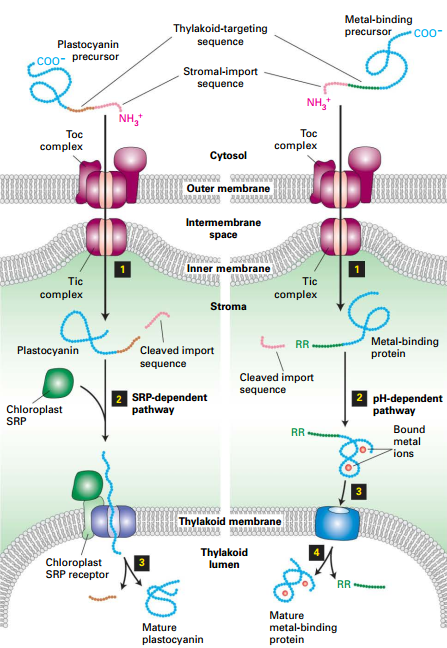
Two Pathways to Chloroplasts Intermembrane Space
- Through Toc|Tic → Stromal import sequence cleavage → Thylakoid targeting sequence exposes.
Path A:
- Thylakoid - targeting sequence binds SRP → SRP receptor and translocon.
- Translocation of protein into thylakoid lumen.
- Thylakoid- targeting sequence-removed.
Path B:
- Twin arginine signal induces Tat (Twin-arginine translocation).
- pH gradient across thylakoid membrane drives translocation. (Bound metal Ion)
- Cleavage of Tat sequence.
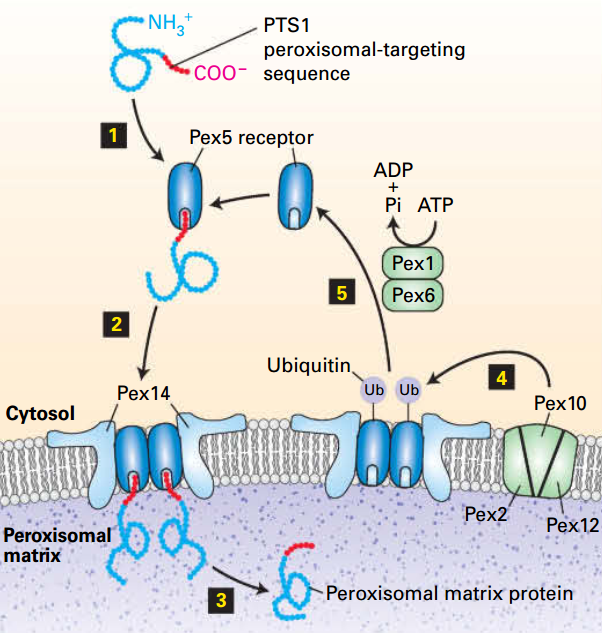
13.5 Targeting of Peroxisomal Proteins
- Luminal proteins are from cytosolic ribosomes, contain a targeting sequence, and are incorporated into the peroxisome post-translationally.
- Folded luminal proteins are imported by a system involving a cytosolic receptor protein and translocation machinery on the peroxisomal membrane.
- Peroxisomal membrane proteins contain different targeting sequences than peroxisomal matrix proteins and are imported by a different pathway.
Targeting sequence (PTS1): C-terminal peroxisomal recognized by Pex5 receptor
Cytosolic Pex5 receptor
Pex14: receptor/channel
Peroxisome luminal proteins:
- Encoded by nuclear genes and synthesized on free ribosomes in the cytosol.
- Incorporated into preexisting or newly generated peroxisomes.
- PTS1 + Pex5 → [PTS1]-[Pex5] → [PTS1]-[Pex5]-[Pex14] → [Pex5]-[Pex14] → PTS1 + Lumen.
- Pex10: [Pex5]-[Pex14] + Ub → Pex14 + [Pex5]-[Ub]→ + ATP → ADP + Pex5 + Ub
- Can be PTS2/Pex7 instead of PTS1/Pex5.