9 Receptor-Mediated Endocytosis|Advanced Cell Biology|Tulane
Receptor-Mediated Endocytosis
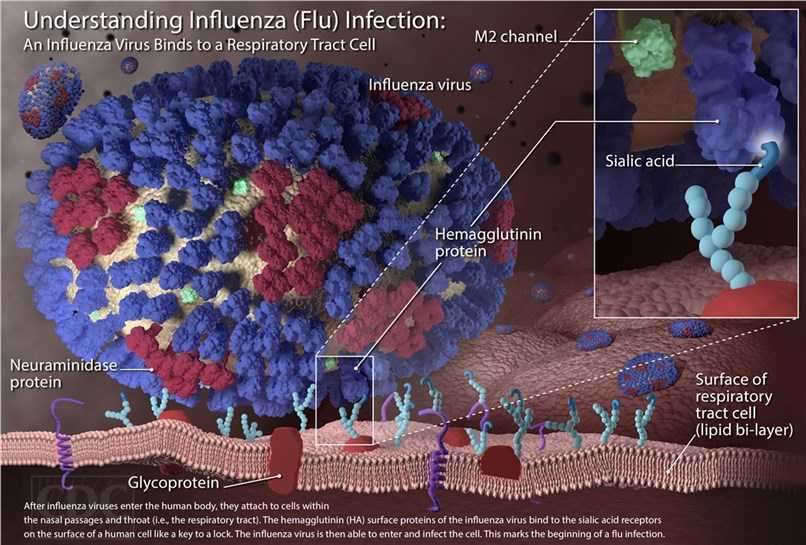
Influenza Virus Infects by Viral Hemagglutinin Interaction with Host Cell Sialic Acid
 |
|---|
| © Creative Biolabs |
Host Antigen: Sialic acid; Viral Antigens: Hemagglutinin (H), Neuraminidase (N)
Neuraminidase cleaves host Sialic acid, enhances release of new viral particles.
Antiviral Neuraminidase inhibitors: Tamiflu (Oseltamivir) and Relenza (Zanamivir). Viral subtypes derive from 18 H and 11 N possible antigens. Vaccine against common H and N antigens. Changes yearly due to antigenic drift.
Endocytosis activate: [Hemagglutinin]-[Sialic]
Lysosome Fuse: H⁺ release to change the pH → Hemagglutinin conformation changed
Neuraminidase: Cleave Sialic → Enhance releases
Antiviral Neuraminidase inhibitors: Tamiflu (Oseltamivir) and Relenza (Zanamivir).
| © Dr.G Bhanu Prakash Animated Medical Videos; 2019 youtube |
14.5 Receptor-Mediated Endocytosis
- Clathrin-coated vesicles: [Ligands]::[AP2-targeting sequences]-[Receptors]
- Ligands delivers: Exp: LDL
- Dissociation: Receptor-ligand complexes dissociated in acid environment:
- receptor → cytoplasm → recycling
- ligand → lysosome → degradation
- Ttransferrin carrier: Iron endocytosis Fe3+ → endosome and recycle transferrin
• Extracellular ligands bound to specific cell-surface receptors with cytoplasmic domain AP2-targeting sequences are internalized by clathrin-coated vesicles.
• The endocytic pathway delivers some ligands (e.g., LDL particles) to lysosomes, where they are degraded.
• The late endosome acidic environment dissociates most receptor-ligand complexes for receptor recycling to the plasma membrane and ligand degradation in lysosomes.
• The iron endocytosis pathway releases Fe3+ in the late endosome but recycles the transferrin carrier proteins with the receptor to the plasma membrane.
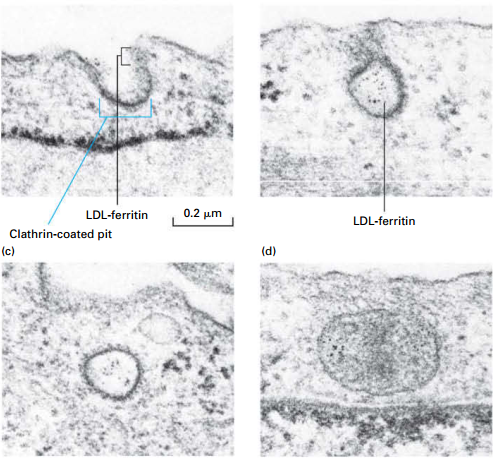
Initial Stages of RME of Low-Density Lipoprotein (LDL) Particles
 |
| © iotech Review; 2014 youtube |
RME: Receptor-mediated endocytosis
LDL: low-density lipoprotein
- RME is used by cells to import specific macromolecules or complexes too large to be imported by membrane transporters
- Uptake specificity is receptor-dependent, in clathrin/AP2-coated vesicles.
Experiment:
- LDL particles (contain cholesterol): labeled with electron-dense, iron-containing ferritin protein (visible with EM)
- Cultured fibroblasts incubated with LDL-ferretin at 4°: LDL binds to LDL-receptors; endocytosis process is inhibited
- Cells warmed to initiate endocytosis.
RME receptors:
- Some types cluster in clathrin-coated pits by cytoplasmic domain association with AP2 even in absence of ligand.
- Others types diffuse freely in the plasma membrane until a ligand-induced conformational change associates them with AP2.
- Two or more types of receptor-bound ligands, such as LDL and transferrin, can be present in the same coated pit/vesicle.
Cellular Uptake / Degradation of LDL by RME
 |
|---|
| © Lodish, Molecular Cell Biology, Eight Edition, p661 |
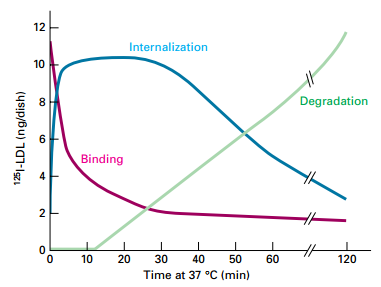
• LDL-LDL receptor complexes completely bound at membrane surface in AP2-clathrincoated pit.
• Internalization of LDL-LDL receptor receptor increases as binding at cell surface decreases.
• Degradation begins within 10-15 minutes, as internalization reached its maximum.
Pulse-chase experiment demonstrates precursor-product relations in cellular uptake of LDL.
Experiment:
- LDL labeled with 125I
- Cultured normal human skin fibroblasts – incubated with 125I-LDL for 2 hours at 4°C (pulse) – LDL binds to surface LDL receptors; not endocytosed
- Unbound LDL – washed away
- Shift to 37°C – activates RME (chase)
- Results:
- 125I-LDL rapidly disappears from surface (binding) by RME.
- 125I-LDL in internal vesicles rises coincidently.
- After a 15 min lag – 125I-LDL degradation in lysosomes increases.
Model of LDL Particle
 |
|---|
| © Lodish, Molecular Cell Biology, Eight Edition, p661 |
• Cells take up lipids from the blood in the form of large, welldefined lipoprotein complexes (LDL, VLDL, HDL, Chylomicon).
• All classes of lipoproteins have the same general structure. Model of LDL Receptor and LDL Binding
• Shell composed of a phospholipid monolayer (not bilayer) containing cholesterol. Interacts with aqueous environment for transport in blood.
• Apolipoprotein B (ApoB) is ligand for LDL receptor.
• Hydrophobic core is mostly cholesteryl esters/triglycerides (minor neutral lipids [vitamins]).
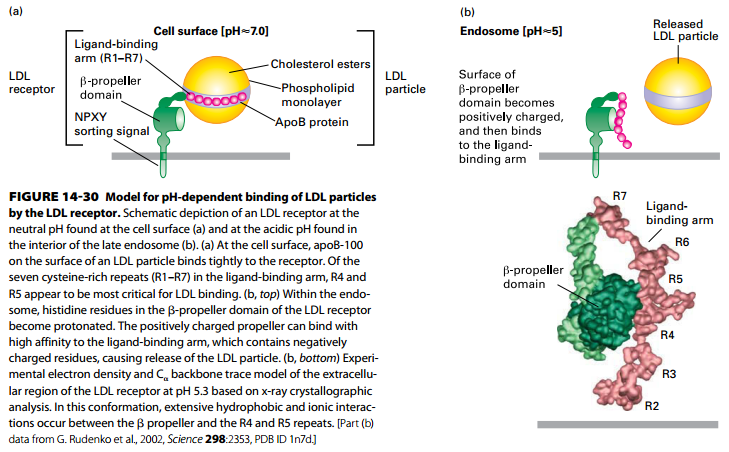
Model of LDL Receptor and LDL Binding
 |
|---|
| © Lodish, Molecular Cell Biology, Eight Edition, p663 |
(a) LDL receptor at neutral pH (binding LDL at cell surface):
- Ligand binding arm seven cysteine-rich repeats (R1–R7) – tightly bind LDL apoB-100 (R4 and R5 – most critical for LDL binding)
- Note receptor NPXY AP2- targeting cytosolic domain.
(b) LDL receptor at acidic pH (releasing LDL in endosome):
- β-propeller domain histidine residues become protonated.
- Positively charged propeller domain binds negatively charged ligand-binding domain → release of LDL particle.
PS: (b, bottom) LDL receptor at pH 5.3 structure – extensive hydrophobic and ionic interactions between the β propeller and R4 and R5 repeats
- Human disease – Familial Hypercholesterolemia
- Excessive circulating LDL causes cardiovascular disease.
- LDL receptor mutations cause too little LDL uptake/clearance into cells (liver cells).
- Several LDL receptor mutations including a single amino acid mutation in NPXY targeting sequence causes inefficient LDL receptor RME – FH disease.
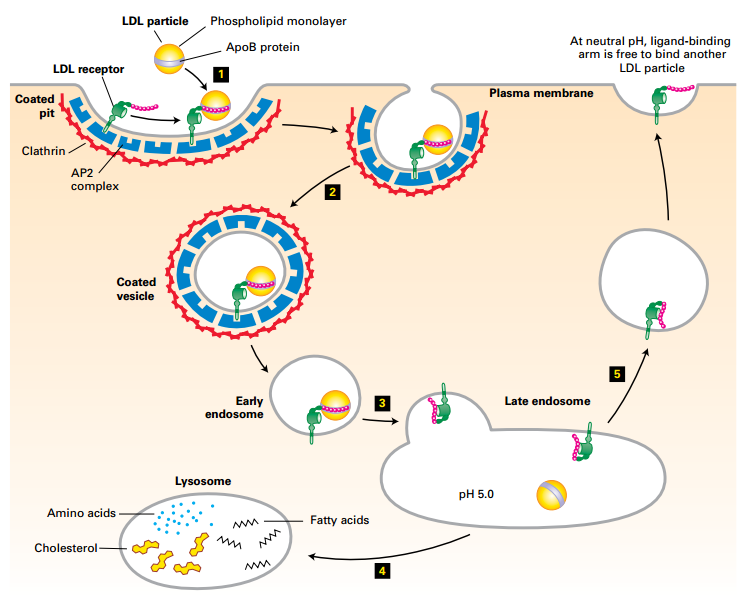
LDL-LDL Receptor Mediated Endocytosis
 |
|---|
| © Lodish, Molecular Cell Biology, Eight Edition, p662 |
Physiologic pH: LDL receptor has high affinity for LDL.
Lysosomal pH: LDL receptor has low affinity for LDL.
Receptor recycled
Clinical Implications: Familial Hypercholesterolemia
- Excessive circulating LDL causes cardiovascular disease.
a. Heterozygous – LDL ~2X elevated in blood
b. Homozygous – LDL 4X-6X elevated
c. Atherosclerotic plaques
b. Premature heart attacks (as early as 20s) - LDL receptor mutations cause too little LDL uptake or clearance into cells (liver cells).
a. LDL receptors are absent
b. Defective LDL binding sites
c. Improper folding → premature receptor degradation
d. NPXY mutation affects sorting to AP2 vesicles
e. Mutation in ApoB ligand, ↓ affinity for LDL receptor - Cells regulate their cholesterol by
a. Inhibition of cholesterol synthesis
b. Down-regulation of LDL receptor
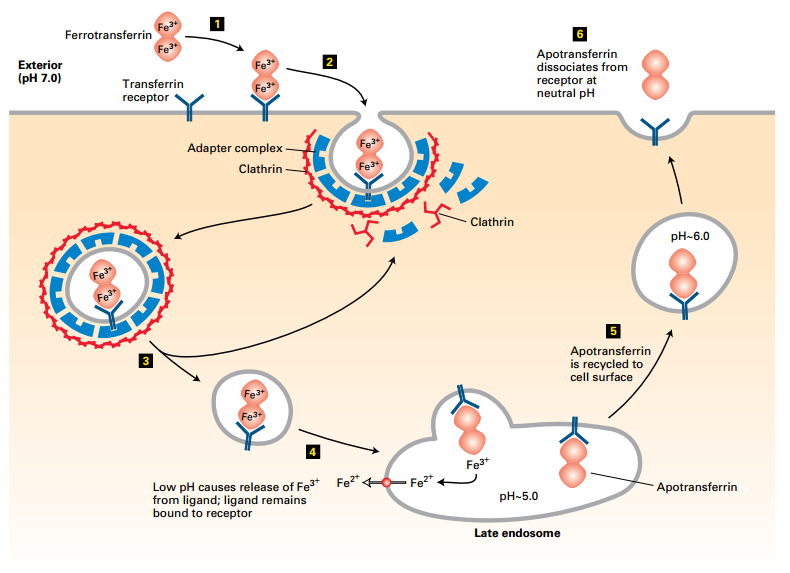
Transferrin Cycle
 |
|---|
| © Lodish, Molecular Cell Biology, Eight Edition, p664 |
-
Transferrin Cycle:
- Ferrotransferrin binds transferrin receptor at cell surface pH 7.0 (1).
- Transferrin receptor forms AP2 clathrin-coated vesicle → RME to late endosome (2-4).
- Acidic pH dissociates Fe3+ → Apotransferrin recycled to cell surface.
- Extracellular neutral pH, receptor releases apotransferrin.
- Fe3+ reduced to Fe2+ for use.
-
Transferrin protein comes in two forms, apotransferrin (not bound to Fe3+) and Ferrotransferrin (carries Fe3+ in blood).
-
Transferrin binding to transferrin receptor is pH-dependent:
- Receptor binds Ferrotransferrin at physiologic pH (pH ~7.0).
- Receptor binds Apotransferrin at lysosomal pH (pH ~5.0).
RME endocytic pathway delivers iron to cells without dissociation of the transferrin–transferrin receptor complex in endosomes.
Transferrin protein:
- Apotransferrin – no bound Fe3+
- Ferrotransferrin – carries Fe3+ in blood
- Binds to transferrin receptor
Uptake mechanism:
- Ferrotransferrin dimer with two Fe3+ binds tightly to transferrin receptor at the cell surface – ~pH 7.0.
- Transferrin receptor cytoplasmic tail interaction with an AP2 adapter complex – incorporates receptor-ligand complex into endocytic clathrin-coated vesicles
- Clathrin disassembly uncoats vesicle.
- Vesicle fuses with late endosome – acidic pH – Dissociates Fe3+ from ferrotransferrin – Fe3+ reduced to Fe2+ by endosome reductase Transported from endosome to cytoplasm Maintains apotransferrin-receptor complex
- Recycling of receptor-apotransferrin complex to the plasma membrane
- Extracellular neutral pH destabilizes complex – releases apotransferrin from receptor
14.6 Directing Membrane Proteins and Cytosolic Materials to the Lysosome
ESCRT (endosomal sorting complex required for transport)
- Endocytosed membrane proteins targeted for degradation in the lysosome are incorporated into vesicles that bud into the interior of the endosome.
- Cellular components (e.g., ESCRT) that mediate endosome membrane budding are used to pinch off enveloped viruses such as HIV from the plasma membrane of virus-infected cells.
- Autophagy envelopes a region of cytoplasm or an organelle into a double-membrane autophagosome for delivery to a lysosome.
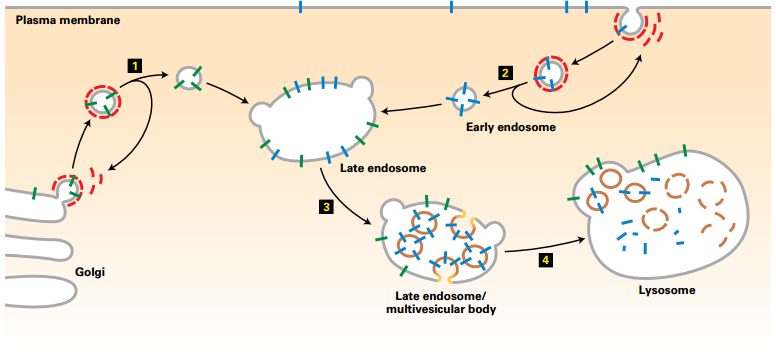
Delivery of Plasma-Membrane Proteins to Lysosome Interior for Degradation
Membrane proteins for degradation → delivered to lysosome lumen
 |
|---|
| © Lodish, Molecular Cell Biology, Eight Edition, p666 |
Mechanism:
- Vesicles carrying newly synthesized lysosomal membrane proteins (green) from the trans-Golgi network – fuse with the late endosome
- Endosomes carrying endocytosed plasma-membrane proteins (blue) targeted for degradation – fuse with the late endosome
- Late endosome:
- Plasma membrane proteins targeted for degradation – incorporated into vesicles that bud into the interior of the late endosome
- Forms a multivesicular endosome
- Multivesicular endosome fuses with a lysosome:
- Internal vesicles (containing targeted membrane proteins)– degraded
- Lysosomal membrane proteins – not degraded
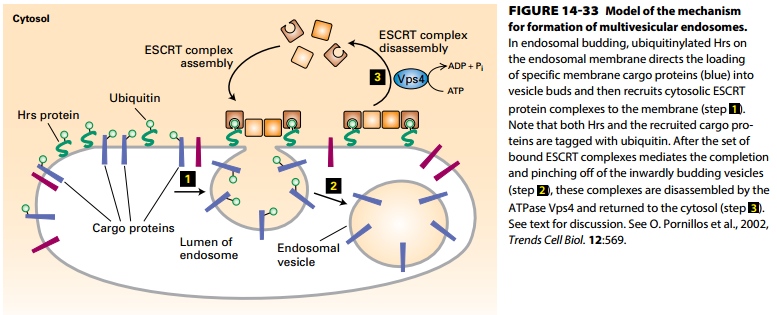
Formation of Multivesicular Endosomes
 |
|---|
| © Lodish, Molecular Cell Biology, Eight Edition, p667 |
- Proteins for multivesicular endosome degradation are tagged with ubiquitin at the plasma membrane, in the trans-Golgi network, or in the endosomal membrane (cargo proteins).
- Ubiquitinated Hrs protein on endosomal membrane direct loading of ubiquitinated membrane cargo proteins (blue) into multivesicular endosomes.
- Cytosolic ESCRT protein complexes and Hrs mediate pinching off of inwardly budding vesicles.
- Vps4 ATP hydrolysis drives disassembly and recycling of ESCRT complex proteins.
HIV Budding from Plasma Membrane
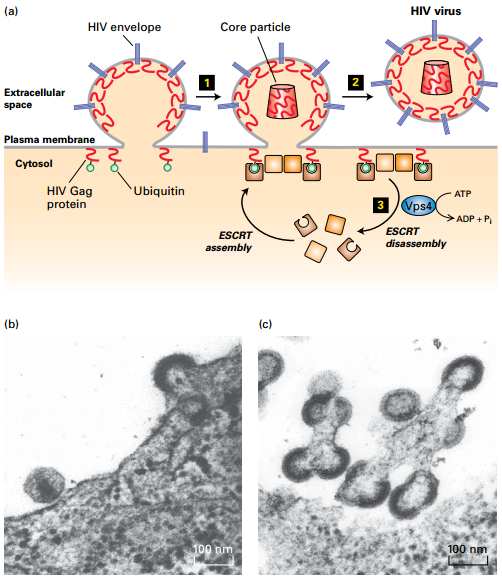 |
|---|
| © Lodish, Molecular Cell Biology, Eight Edition, p668 |
• Retrovirus (HIV) budding from plasma membrane exploits ESCRT/Vps4 machinery for multivesicular endosomes.
• Ubiquitinated viral Gag proteins function like Hrs protein → recruit ESCRT/Vps4 complexes to pinch off viral particle. The Autophagic Pathway
• Autophagic pathway delivers cytosolic proteins and organelles to lysosomes for degradation.
• ATG proteins induce formation of cup-shaped membrane structure around a portion of the cytosol (right) or an organelle (left) to create am autophagosome.
• Autophagosome envelop contents in two complete membranes.
• Fusion of autophagosome outer membrane with the lysosome membrane releases a singlemembrane vesicle and its contents into the lysosome interior for degradation.
Mechanism:
ATG proteins induce formation of cup-shaped membrane structure around
- (right) a portion of the cytosol
- (left) an organelle such as a mitochondrion
- Proteins involved include Atg 5, 8, 12, 16 – Atg 8 forms coat around autophagosome
Step 1: Continued membrane addition and fusion forms autophagosome – envelops contents in two complete membranes
Step 2: Fusion of the autophagosome outer membrane with the lysosome membrane –
- Releases a single-membrane vesicle and its contents into the lysosome interior
- Vesicle protein and lipid components – degraded by lysosomal hydrolases
- Amino acids – permease transport across the lysosomal membrane into the cytosol for use in protein synthesis
9 Receptor-Mediated Endocytosis|Advanced Cell Biology|Tulane
https://karobben.github.io/2021/10/18/LearnNotes/tulane-cellbio-9/








