10 Cell Signaling|Advanced Cell Biology|Tulane
Cell’s social network
No cell lives in isolation; life requires that all cells sense chemical and physical stimuli in their environment and respond which changes that can affect their function os development.
– Molecule Cell Biology, 3ed
| Inner signals | Toxic signals | Physical signals | Responding to other biologies |
|---|---|---|---|
Responding of the cell signaling:
- differentiation
- proliferation
- exocytosis
- migration
- apoptosis
- adhesion
- senescence
Receptors:
- Three domains:
- Extracellular domain
- plasma-membrane-spanning domain
- intracellular domain
- Acts like a ligand
- Many signaling proteins are belongs to signal transduction proteins
Transmembrane signaling: only a few mechanisms
- Ligand-gated ion channel (iontropic receptros)
- G-protein-coupled receptors (metabotropic)
- Kinases-linked receptors
- Nuclear receptors
Signal Transduction
Snthesis → Release → Transport → Detection → Initiation → Functional Change → Deactivation → Removal
- Synthesis of the signal
- Release of the signaling molecule by the signaling cell: exocytosis, diffusion, cell-cell contact
- Transport of the signal to the target cell
- Detection of the signal by a specific receptor protein
- Initiation of one or more intracellular signaltransduction pathways
- A change in cellular metabolism, function or development triggered by the receptor-signal complex
- Deactivation of receptor
- Removal of the signal (downregulation)
Signaling events are ordered both spatially and temporally
Cellular Tools for information transmission
- Ligands
transmit signals - Receptors
receive information in form of a ligand transmits signal across membrane - Transducers
pass information enzymatically active may be signal integrators - Adapters
no catalytic activity modulate proximity of transducers - Scaffolds
provide architecture allow energetically unfavorable events - Effectors
perform an end function
Ligands (structure)
Chamical structure, small molecules:
• Small molecules (e.g. amino acid or lipid derivatives, acetylcholine)
• Peptides (e.g. ACTH, vasopressin)
• Steroids
• Retinoids
• Thyroxine
• Proteins (usually large & hydrophilic, bind to cell-surface receptors)
(hydrophobic, bind intracellular receptors)
Major classes
• Hormones
• Growth factors, cytokines, chemokines
• Neurotransmitters
• Pheromones
• Can also be changes in metabolite concentration, e.g. oxygen or nutrients or physical stimuli such as light and heat
Models of Cell Signaling
- ENDOCRINE SIGNALING
Example: release of insulin by cells in the pancreas, travels in the blood stream and acts on distal liver, muscle and fat cells - PARACRINE SIGNALING
Examples: Growth factors and cytokines that signal to neighboring or surrounding cells - AUTOCRINE SIGNALING
Example: PDGF binds to PDGFproducing and secreting cells to stimulate cell growth - SIGNALING BY MEMBRANEATTACHED PROTEINS
Example: Delta-Notch signaling
Not every ligand that binds to a receptor also activates the receptor
- Agonists are able to activate the receptor and result in a maximal biological response. The natural endogenous ligand with the greatest efficacy for a given receptor is by definition a full agonist (100% efficacy).
- Partial agonists do not activate receptors thoroughly, causing responses which are partial compared to those of full agonists.
- Antagonists bind to receptors but do not activate them. This results in receptor blockage, inhibiting the binding of agonists and inverse agonists.
- Inverse agonists reduce the activity of receptors by inhibiting their constitutive activity.
What determines the cellular specificity of responses to ligands?
- The presence or absence of receptors
- The internal signal transduction or response machinery of individual cell types
Ligands induce specific cellular response
One type of ligand could trigger different type of respons in diffrent cell types.
Exp: Acetylcholine could initiate:
- Pancreatic Acinar cell: Digestive enzymes
- Pancreatic β cell: Insuline
- Smooth Muscle: contraction
- Parotid gland (saliavary): amylase enzyme
Receptors
They work as a gateway to the cell. As a result, they are crucial … to regulate almost every known physiological process.” (Robert Lefkowitz)
Intracellular (Nuclear) Receptors
• Can be located in the cytoplasm or the nucleus
• Bind to hydrophobic ligands that can diffuse across the plasma membrane
• Contain DNA-binding domains and act as ligand-regulated transcriptional activators or suppressors
• Characteristic lag period between ligand binding and cellular response of 30 minutes to several hours
• Effects of NR agonists can persist for hours or days after plasma concentration is zero
Cell-surface receptors
Hydrophilic ligands bind to cell-surface receptors
• Integral membrane proteins
• Domain structure
- Extracellular
- Transmembrane
- Intracellular
• Exhibit ligand binding specificity
• Ligand binding induces conformational change that exerts an effect intracellularly
• Effector specificity
Seven Major Classes of Cell-Surface Receptors
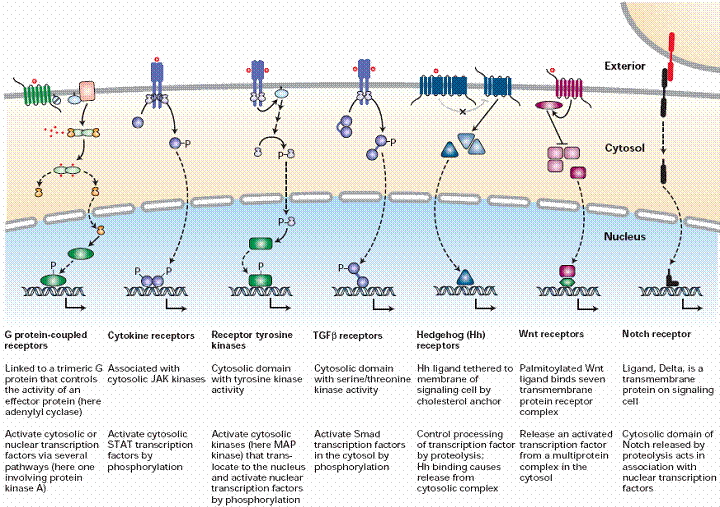 |
|---|
| © Molecular Cell Biology, Edt3 |
Experimental Figure 15.3 Ligand Growth hormone binds to its receptor through molecular complementary. Binding Specificity
-
Ligands binding to different types of receptors lead to different physiological responses
-
Sensitivity of a cell to an external signal is determined by the number of surface receptors
-
Maximal cellular response does not require binding of all receptors
50% of maximal response when only 18% of receptors bound with ligand
80% of maximal response is induced when 50% of the receptors are occupied
Transducers
pass information; enzymatically active; may be signal integrators
Protein kinases and phosphatases are employed in virtually all signaling pathways
- Regulation of protein activity by a kinase/phosphatase switch.
- A simple signal transduction pathway involving one kinase and one target protein.
- ~ 600 kinases and 100 different phosphatases in human genome
- Target residues are ‘phosphoacceptors’ - Ser, Thr or Tyr
- Assembled into the ‘KINOME’
- Diversity encompasses transmembrane, simple and complex structures
Post-translational modifications (PTM)
-
Reversible addition of a small chemical group causes change in activity or location of a signaling protein
-
PTMs require the action of both modifying and unmodifying enzymes (allowing the signal to be given and terminated)
-
GTPase switch proteins cycle between active and inactive forms.
-
Switching mechanism of G proteins.
Second messengers amplify the signal
Second messengers
- Short lived, diffusible intracellular signaling molecules
- Elevated concentration leads to rapid alteration in the activity of one or more cellular enzymes
- Removal or degradation terminates the cellular response
Adapters and Scaffolds
no catalytic activity modulate proximity of transducers
provide architecture allow energetically unfavorable events
Signaling induced by protein-protein interactions
Cellular Tools for information transmission
Adapter has no catalytic activity, modulate proximity of transducers
Many signal-transduction pathways contain large multiprotein signaling complexes which are held together by adapter proteins
- These regulatory interactions are mediated by specific protein domains
Signaling proteins are modular, consisting of groupings of highly conserved domains each with a specific function.
When used in combination, these domains allow the construction of specialized molecules with multiple input and output points. Use of domains ensures function only in the correct conditions/context
Adapter - Protein clustering
Clustering of neurotransmitter receptors in the region of the postsynaptic plasma membrane adjacent to the presynaptic cell promotes rapid and efficient signal transmission
- PDZ domains are protein-protein interaction domains recognizing mainly the C-termini of their target proteins
- Src-homology 2 (SH2) domains bind to specific phospho-tyrosine containing peptide motifs.
- Src-homology 3 (SH3) domains bind to proline rich peptides
- Pleckstrin homology (PH) domain Bind to phosphoinositides, bg-subunits of G-proteins and PKC
Scaffolding protein provide architecture and allow energetically unfavorable events
Lipid Rafts
Rafts, 10-50 nm, contain a max of 50 proteins along cholesterol, sphingolipids, and Glycosylphosphatidylinositol-anchored proteins
Caveolae are a special type of lipid raft
Coalesce with active signaling and might concentrate signaling proteins
May provide a microenvironment for signaling and help GPI-linked proteins signal across membrane
Caveolae are small (50 -100 nm) invaginations in plasma membrane containing
Caveolin on the cytosolic leaflet of the plasma membrane
Caveolin helps form the flask-shaped pits involved in endocytosis
Caveolin interacts with cholesterol and may be play a role in transfer of cholesterol into lipid raft domains.
Signal proteins that attach to the plasma membrane via lipid anchors tend to be concentrated in caveolae.
Effectors
perform an end function
Cellular Tools for information transmission
Many ligands bind to multiple types of receptors leading to different physiological responses
- Different Receptor ligand complexes can activate the SAME response:
Example: Epinephrine or glucagon can activate glycogen breakdown and release of glucose into the blood - Turning off or dampening signaling
Signaling Pathways often cross-communicate Signaling Networks
the same cellular response may be induced by multiple signaling pathways by distinct mechanisms
Interaction of different signaling pathways permits fine-tuning of cellular activities
Cell-Cell Adhesion and Communication- Integrating Cells into Tissues
Cell adhesion molecules (CAMs) classes:
- cadherins – cell-cell adhesion; calcium dependent e.g, E-cadherin, P-cadherin
- Ig superfamily of CAMs – cell-cell adhesion; calcium-independent; some are found enriched on specific cell types – e.g. N-CAM, V-CAM
- Integrins – cell-matrix adhesion molecule e.g. a1 integrin, b1 integrin
Signal Transduction
- The effects of activation of cell surface receptors are more complicated than a simple step-by-step cascade
- By no means is signaling a linear event
- Extensive networking and cross talk
- Key is integration
- Why so complicated?
Amplification - Reliability - Redundance
When signaling goes wrong
- Dysregulated signaling results in inappropriate responses to stimuli
- Over- and under-reaction are equally devastating
- All disease is the result of
- inappropriate,
- inadequate or
- over-enthusiastic signal transduction
When the target ignores the Signal:
Losing the Signal → Type I Diabetes
When the target ignores the Signal → Type II diabetes
Too much signal → Stroke
Multiple breakdowns → Cancer
When a signal doesn’t reach its target → Multiple Sclerosis
- Mutations of receptors may lead to constitutive activity
- Amplification or overexpression of Her2/neu is associated with aggressive breast cancers
- Suppression of pro-apoptotic genes will lead to transformation
- Some cancer-causing viruses encode an activated form of src
- 90% human tumors have activating Ras Mutations; Activating mutations of Ras, Rac, Rho or cdc42 are Enough to induce cellular transformation In the lab
Oncogenic transformation
- Uncontrolled Proliferation
- Suppressed Apoptotic
- Downregulation of antigenic surface proteins
Many signaling proteins are oncogenes: activating or inactivating mutations are sufficient to cause transformation
Studying Cell-Surface Receptors and Signal Transduction Proteins
Kd = dissociation constant measures the affinity of the receptor for the ligand
Low Kd = high affinity of ligand for receptor
High Kd = low affinity of ligand for receptor
- For high-affinity ligands, binding assays can determine the Kd and the number of receptors per cell
- Suspension of cells incubated for 1 h at 4oC with increasing conc. of 125I-labeled insulin
- Pellet cells and wash away unbound insulin
- Measure radioactivity = total binding
- Repeat binding assay in the presence of 100-fold excess unlabeled insulin = nonspecific binding
- Subtract nonspecific binding from total binding = specific binding
- Determine total number of receptors per cell
- Determine Kd
Isolation of Membrane receptors
A typical mammalian cell has between 1,000 and 50,000 copies of receptor
Affinity labeling: crosslink radiolabeled ligand to receptor- follow radioactivity in purification
Affinity chromatography: ligand is chemically linked to polystyrene beads - pass homogenate over column - receptor binds - release receptor by passing excess ligand through column
Cloning of Receptors
- Allow identification of receptors that constitute small percentage of total cellular protein, and identification of receptors from small tissue source
- Expression cloning
- Homology cloning
Expression Cloning of Receptors
- Make cDNA library from appropriate tissue
- Express library in cell line that does not express the receptor of interest
- Use ligand binding to identify clone that encodes receptor
Functional Expression Assay
- Divide library into smaller pools to find clone that encodes receptor
- Provided cells express relevant signal-transduction proteins, transfected cells will now exhibit normal cellular response to ligand X if the cDNA encodes the functional receptor
Homology Cloning
- find additional variants of receptor
- find novel receptors
- Degenerative PCR (odorant receptors)
- Or screen library at low stringency (glutamate receptors)
- Or database search (Taste receptors)
Degenerative PCR Cloning
• Design degenerate primers that match amino acid sequences in conserved region of receptor family members
• Use these primers in all pairwise combinations to amplify related sequences in cDNA prepared from tissue suspected of expressing the new receptor
• Clone and sequence DNAs from PCR
• Examine sequence for hallmarks of receptor family
• Use these DNAs as probes to screen cDNA library from tissue known to express receptor
• Examine proteins encoded by positive cDNA clones
The structures and actions of receptors may be studied by using biophysical methods such as X-ray crystallography, NMR, circular dichroism, and dual polarization interferometry.
Computer simulations of the dynamic behavior of receptors has been used to gain understanding of their mechanism of action.
10 Cell Signaling|Advanced Cell Biology|Tulane
https://karobben.github.io/2021/10/21/LearnNotes/tulane-cellbio-10/









