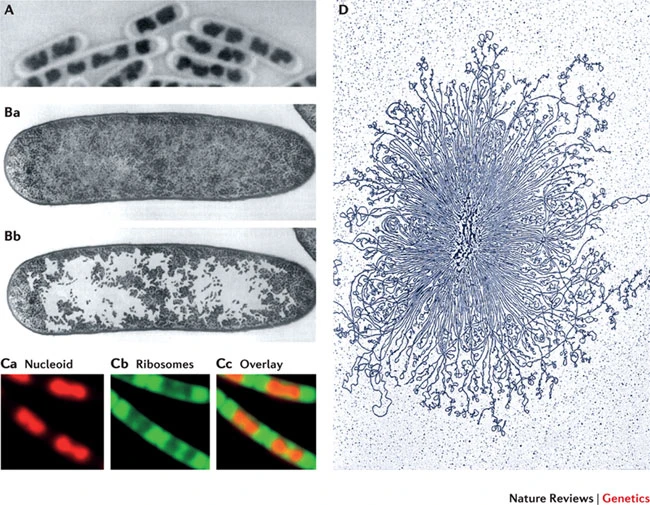
Chromosome Structure
DNA fiber and its packaging
Introduction:
E.coli, is 4.6 million base pairs (approximately 1.1 mm, if cut and stretched out (0.5 uM in width and 2 uM in length)
Each human cells contains several meters of DNA (3.2 x 10 9 nucleosides ) if stretched end-to-end, the nucleus of a human cell, which contains the DNA, is only about 6 μm in diameter. This is geometrically equivalent to packing 40 km (24 miles) of extremely fine thread into a tennis ball!
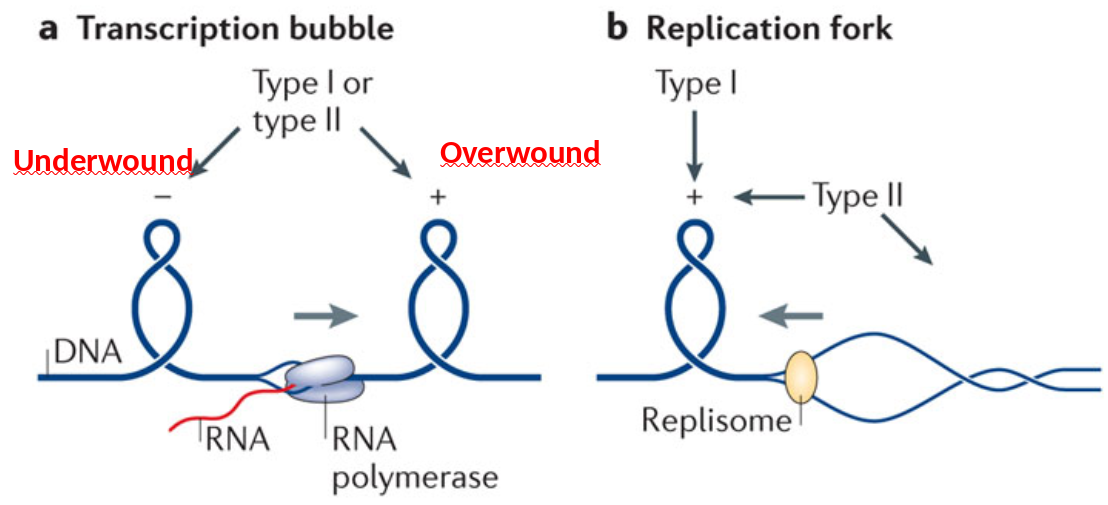
In bacteria, DNA gyrase aids in DNA packaging by causing an accumulation of negatively supercoiled (underwound) DNA.
Some proteins are known to be involved in the supercoiling; other proteins and enzymes such as DNA gyrase (Topoisomerase)help in maintaining the supercoiled structure.
 |
|---|
| © Xindan Wang, 2013 |
Supercoiled DNA
Winding
 |
|---|
| © psu.edu |
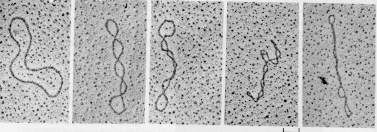
Supercoiled DNA can acquire different conformations or shapes (topologies). Excessively unwound DNA molecules exist as topological isomers with negatively supercoiled (plectonemic or solenoidal) forms.
Unwinding
 |
|---|
| Unwinding a DNA molecule without allowing it to rotate creates supercoils |
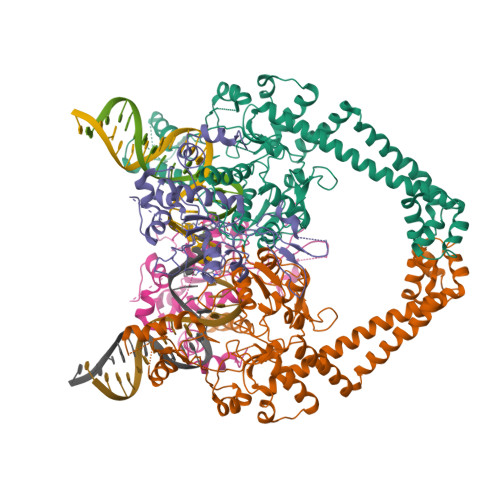
topoisomerase I
Type I topos cleave single strand through use of covalent Tyr-DNA intermediate
- Type IA: relax negative supercoil; Tyr-5’-P-DNA (free 3’-OH)
- Type IB: relax pos. or neg. supercoil; Tyr-3’-DNA (free 5’-OH)
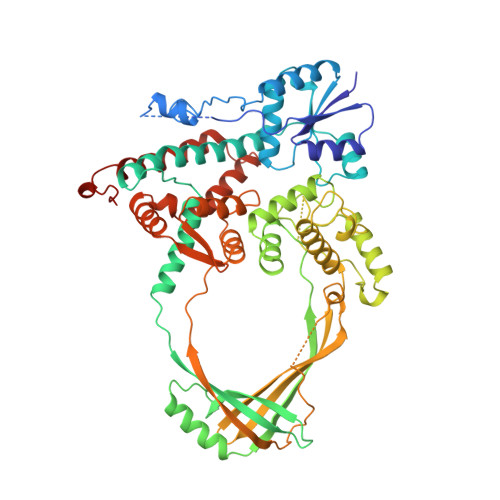 |
 |
|---|---|
| © PDB, Topoisomerase I | © PDB, Topoisomerase II |
| Type I topos cleave single strand through use of covalent Tyr-DNA intermediate |
All Type II topos can relax pos. and neg. supercoils and cleave double strand: DNA gyrase (prokaryotic) is the only enzyme that can introduce neg. supercoils |
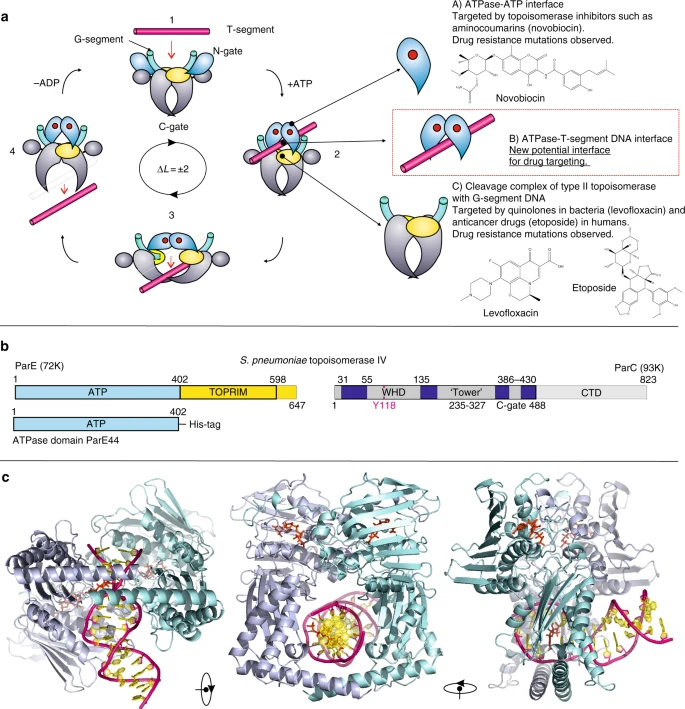 |
|---|
| © Ivan Laponogov, 2018 |
Topoisomerase Inhibitors
Topoisomerase Inhibitors are Useful Anti-biotic & -cancer Therapeutics
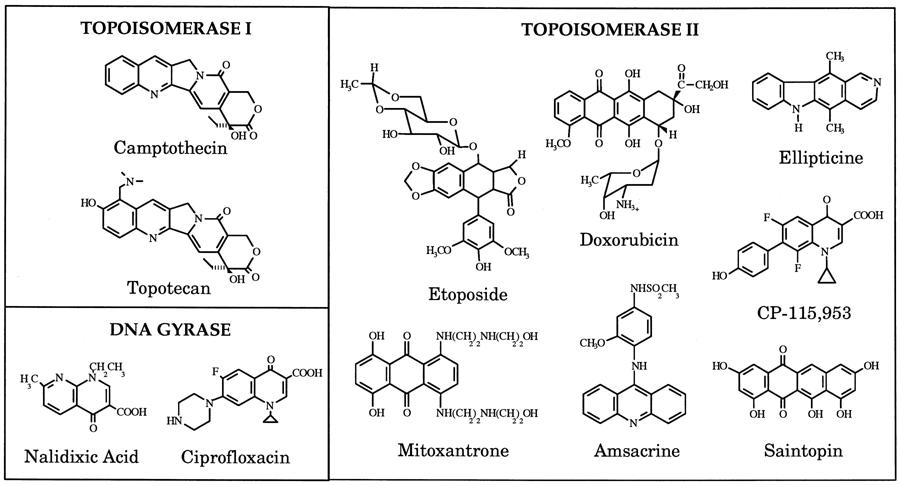 |
|---|
| Unknow Source |
Eukaryotic Chromosome
| © Ronald Hancock, 2012 |
 |
 |
|---|---|
| © lumenlearning | © microbenotes, 2021 |
Compositions:
- DNA
- Histones: H1, H2A, H2B, H3 and H4
- Topoisomerase II
Histon Genes
 |
|---|
| © Izabela Makalowska, 1999 |
Histones Genes:
- No introns
- Multigene compound clusters
- Genes are duplicated (H2A, H2b, H3, H4)
- Highly conserved evolutionarily (except H1)
- Histone variants (CENP-A and H2AZ, etc.)
Histone Core and the Nucleosome
Nucleosome:
- Nucleosome Histone Octamer Core:
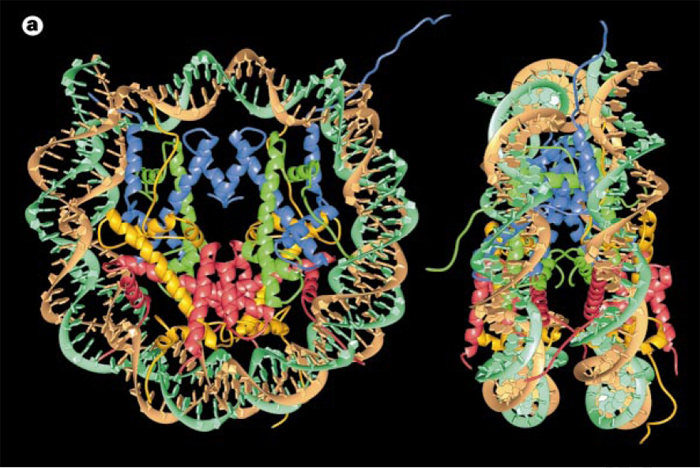
H2A, H2B, H3, H4 - DNA:Protein complex: 8 histone core proteins ~146 bp of wrapped DNA (twice) spacer region (~90 bp)
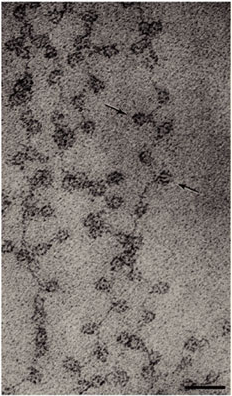
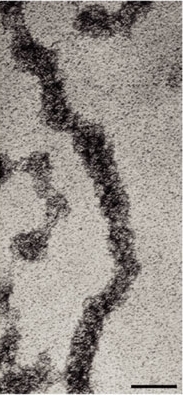
- Forms 11 nm nucleosome chromatin fiber “beads on a string” (~6-7X compaction of DNA).
- Stabilized through electrostatic interactions: DNA phosphate (-) charge Histones (+) charge (Arg, Lys)
DNA wrapped around histones
| © Anthony T. Annunziato, 2008 |
|---|
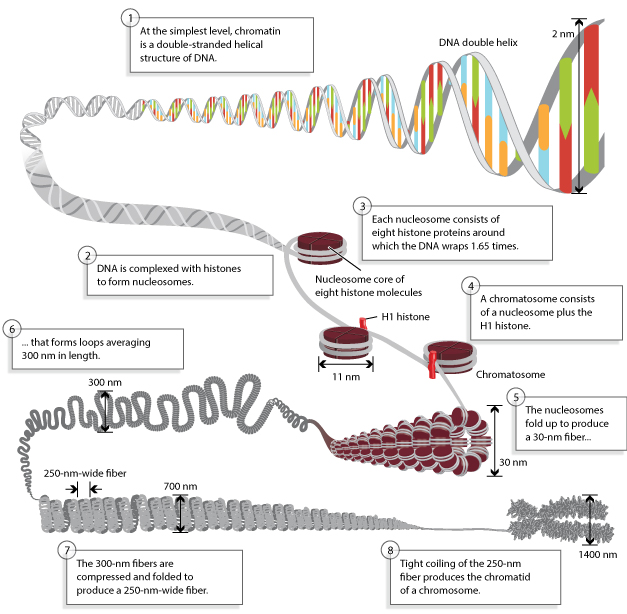 |
 |
 |
 |
 |
|---|
Chromatin Structure
30 nm Chromatin Fiber
- Histone 1 (H1) bind DNA as it wraps nucleosome core.
- Six nucleosomes per helical turn.
- Nucleosomes stacked on top of one another in a zigzag forming (~100X compaction).
- Inactive chromatin in 30 nm fiber (or higher order).
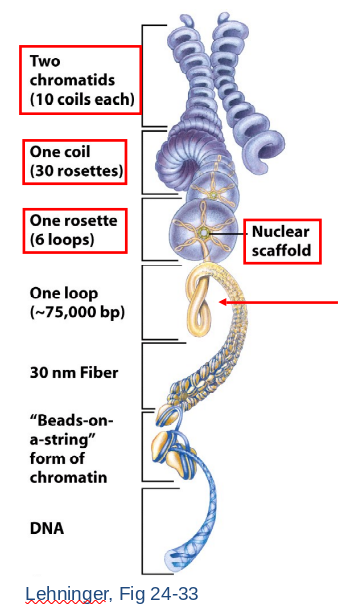
- Higher Order Structure
- 30 nm fiber begins to loop.
- Forms a rosette arrangement constructed upon nuclear scaffold proteins.
- A coil forms of repeated rosettes.
- Each chromatid consists of numerous coils.
However, more evidence suggests a lack of the existence of 30-nm chromatin.
- Fussner, E. et al. Open and closed domains in the mouse genome are configured as 10-nm chromatin fibres. EMBO Rep. 13, 992–996 (2012).
- Nishino, Y. et al. Human mitotic chromosomes consist predominantly of irregularly folded nucleosome fibres without a 30-nm chromatin structure. EMBO J. 31, 1644–1653 (2012).
- Hansen, J. C. et al. The 10-nm chromatin fiber and its relationship to interphase chromosome organization. Biochem. Soc. Trans. 46, 67–76 (2018).
Instead, 11-nm fibers form DNA-loops
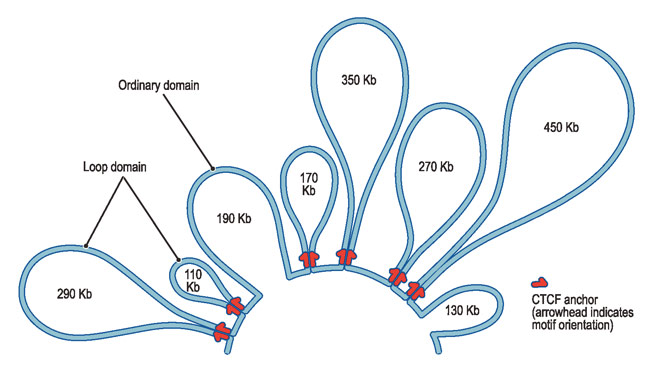 |
|---|
| © harvard.edu |
Interphase Chromatin
 |
|---|
| © David Saintillan, 2018 |
-
Euchromatin (lightly staining)
- 11 nm active chromatin.
- Gene expression “on”.
- DNA replication of S phase.
-
Heterochromatin (darkly staining)
- Condensed, inactive chromatin.
- Gene expression “off”
Q&A
Q: How to read genetic information from highly packed chromatin?
A: Basic principle: Loosen packed chromatin
- Post-translational modification on histone tails (enzymes)
- Chromatin remodeling (chromatin remodeling complex)
Histone Tails Modifications
 |
|---|
| © Jochen Erler, 2014 |
Occur on exposed histone tails (dashed lines).
3 Types of Modifications :
Acetylation: decondensation.
Methylation: condensation → prevents acetylation.
Phosphorylation: decondensation → creates (-) charge. Oddly H3Ser10 phos. → condensation.
Acetylation: Neutralizes histone (+) charge and electrostatic attraction to DNA.
Opens chromatin → beads on string.
HATs – Histone acetyltransferases on lysines e- amino groups (H3lys9).
Deacetylation: Maintains (+) charge and electrostatic attraction to DNA.
Closes chromatin – 30 nm fiber
HDACs – Histone deacetylases
Constituitively assoc. with silent genes
Chromatin less sensitive to DNAse.
Regulation: Chromatin Unwinding/Winding
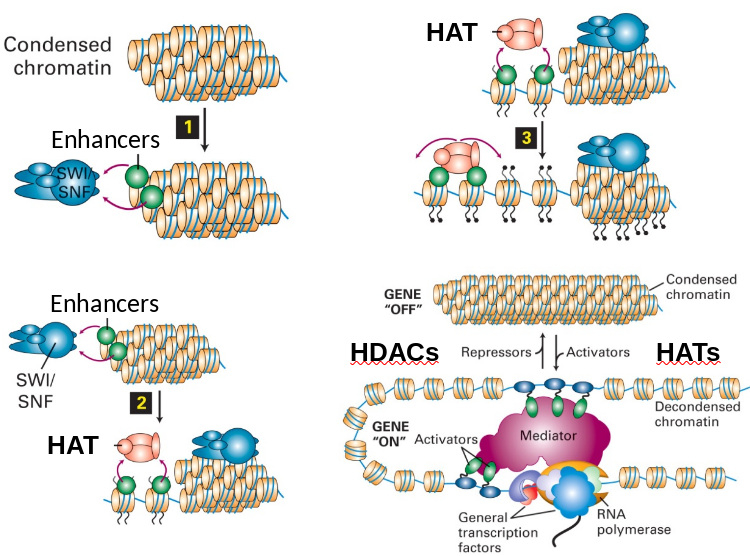 |
|---|
| Unknow Source |
Activators (SWI/SNF) and repressors (H1) promote unwinding or winding.
- SWI/SNF complex binds enhancers and begin unwinding.
- Attracts HATs to acetylate histone tails.
- Further action continues to open chromatin.
- Equilibrium balance between HATs and HDACs to maintain unwinding/winding.
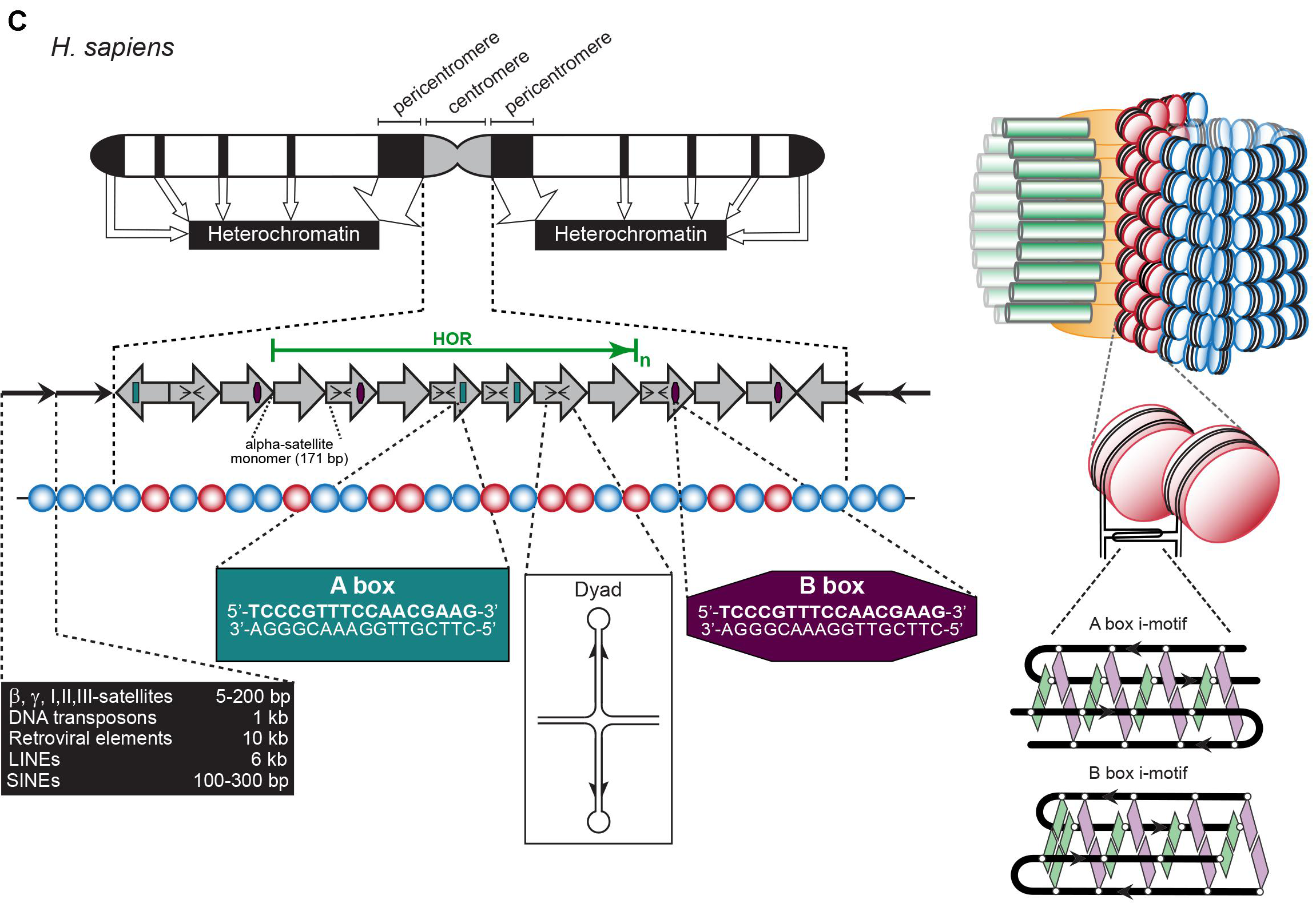
Centromere and Telomere
Telomere: TTAGGG Minisatellite
Centromere: Various satellite components
Hypervariable: ministatellites
Structures see previous picture
- 0.1-20 kb of 6-64 bp repeated units.
- Several MB in length of tandemly repeated 5-170 bp sequences.
- <100 bp repeats dispersed in chromosomes.
- Also Megasatellite: 100s of 3-5 kb repeats at different locations of some chromosomes.
Genome-Wide Repeat Sequences: Transposons
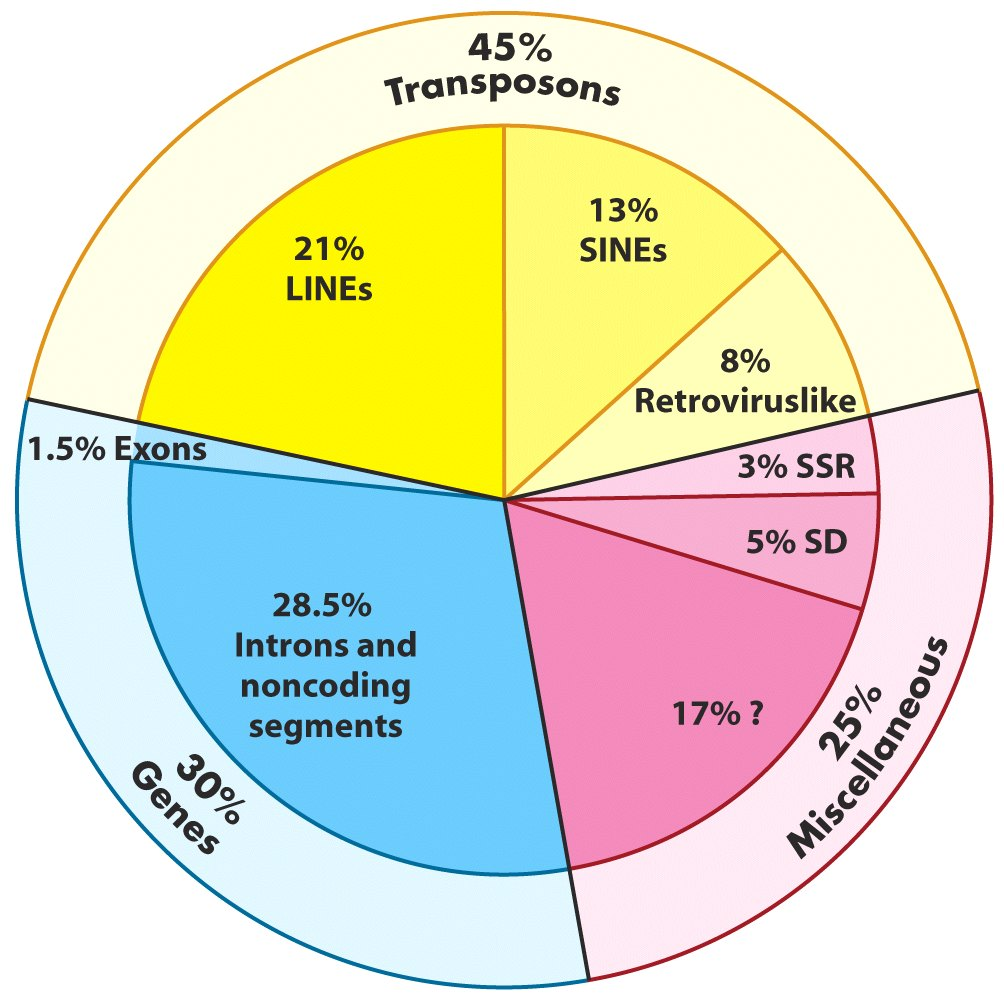 |
|---|
| Lehninger, Fig 24-8 |
- Retrotransposons: replicative (or copy) transposition. LINEs, SINEs and LTRs.
- DNA transposons: Conservative transposition. Cut and paste mechanism.
Autonomous vs. nonautonomous transposition.
Telomeres and The End Replication Problem
- Problem? Lagging strand end will shorten by ~ 1 primer length every genome duplication
- Solution? Telomerase!
Ribonucleoprotein Complex That Catalyzes 3’ Telomere Extension
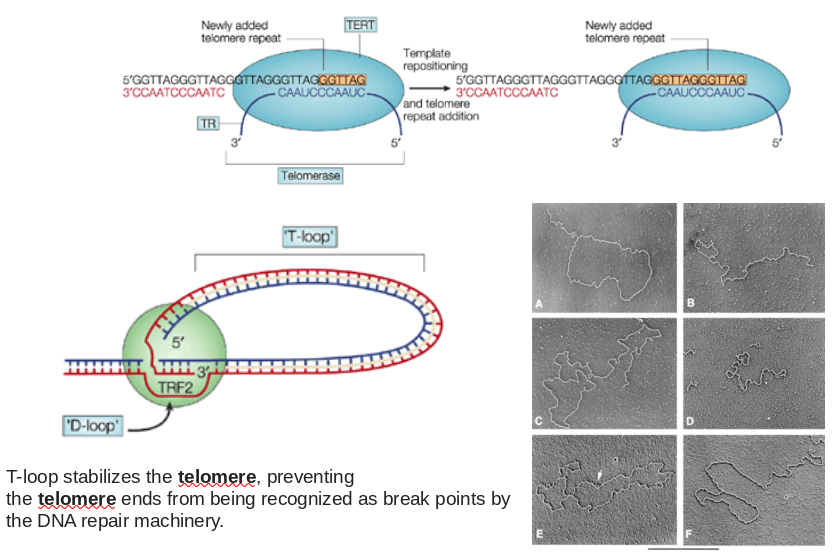 |
|---|
| Source Unknow |
Telomere functions, cancer and aging
- The primary role of telomeres is to protect chromosome ends from recombination, fusion, and from being recognized as damaged DNA.
- Maintaining Telomere length by telomerase is crucial for the survival of cancer cells in the vast majority of tumors. (Highly expressing telomerase can immortalize normal cells)
- Telomere length shortens with age. Progressive shortening of telomeres leads to senescence and apoptosis, affecting the health and lifespan of an individual.
Centromeres across species
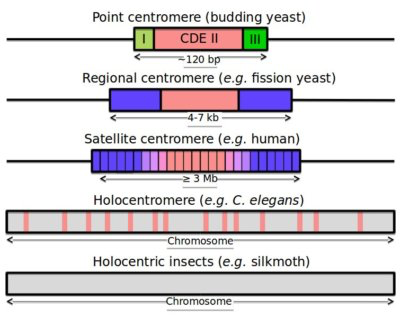 |
|---|
 |
| Source Unknow |
Functions of centromeres and kinetochores during mitosis: chromosome segregation
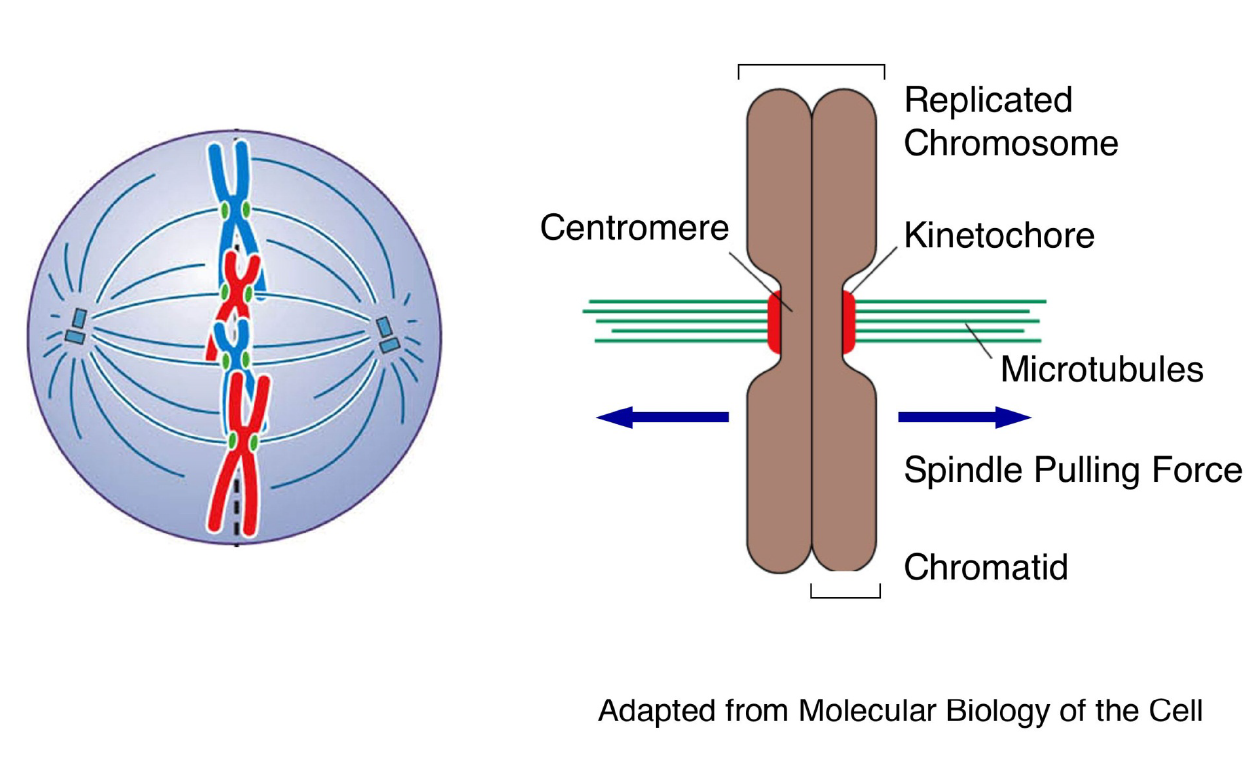 |
|---|
Genomic Instability and Cancer
 |
|---|
| © myeloma.uams.edu |
Chromosome Structure
https://karobben.github.io/2021/11/26/LearnNotes/tulane-biochem-14/








