DNA Transcription, Transcription Factor Binding
RNA polymerases (RNAPs)
- Transcription of DNA to RNA
- RNA is composed of nucleotides containing ribose sugar and four bases uracil, adenine, guanine and cytosine
- Bacteria: One enzyme for all of RNA synthesis
- Eukaryotes: At least three major enzymes (RNAP-I,-II,-III)
RNA Strand Terminology
These terms describe the same strand…
- Template strand – used by RNA pol II to create RNA. Has complementary antiparallel sequence from RNA.
- Non-coding strand – This strand doesn’t code for protein. The complementary antiparallel strand would.
- Antisense strand – This strand is the opposite of the coding strand and thus is antisense.
The RNA message must make sense. Replace T with U.
The eleongation of the RNA is from 5’ → 3’, as the result, the reading direaction is from 3’ → 5’
Bacterial Promoter Consensus Sequences
Expression controlled by interactions of promoter elements with RNA polymerase and specific repressors/activators.
Bacterial Promoter Elements:
- UP element (-40 to -60; A-T rich) strongly stimulates transcription.
- TATAAT (-10) and TTGACAT (-35) regions. Help define transcription start and strand usage.
Bacterial Transcription Initiation
 |
|---|
| © archive.cnx.org |
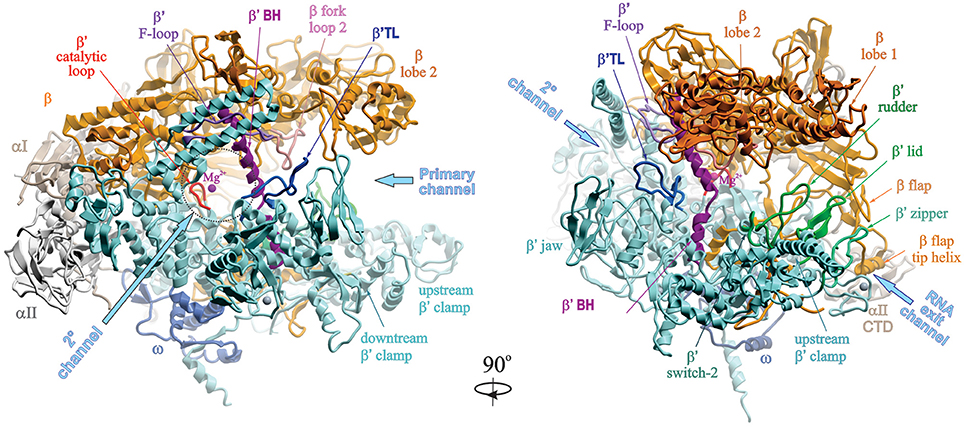
Bacterial RNAP
 |
|---|
| © Jookyung Lee |
- Holoenzyme versus core enzyme.
- Core subunits: α2ββ’ω (400 kDa).
- Holoenzyme: core subunits plus σ factors
- A shape of crab claw formed by the β and β’ subunits
RNAP structure from Thermus aquaticus
Different σ factors of E. coli RNAP
Five σ(Sigma) factors in E. coli gene expression
σ 70: Most genes
σ 32: Heat shock proteins
σ 28: Flagellar operon
σ 38: Stress-response genes
σ 54: Nitrogen metabolism genes
Regulating the transcription of coordinately expressed groups of special-purpose genes
Bacterial Transcription Elongation
During elongation, the prokaryotic RNA polymerase tracks along the DNA template, synthesizes mRNA in the 5’ to 3’ direction, and unwinds and rewinds the DNA as it is read.
 |
|---|
| © archive.cnx.org |
RNA synthesis proceeds in the 5’ -> 3’ direction.
Transcription elongation causing DNA supercoiling, relaxed by topoisomerases
RNAP following the template strand, the transcript being wrapped around the helix every 10bp (no strain and no supercoiling of the DNA).
Instead, DNA rotates avoiding transcript entanglement.
Bacterial Transcription Termination
Intrinsic termination
Transcription termination sites in E. coli
- A series of 4-10 A-T base pairs with A’s on the template strand, transcribed into a poly-U tail at the 3’ end of RNA.
- A G-C rich region with a palindromic sequence that immediately precedes the A-T’s, allowing self-complementary “hairpin” structure formation for the RNA transcript.
- Key factors for proper chain termination
- Structural stability of RNA transcript due to G-C rich hairpin
- Weak base pairing of oligo(U) to template DNA
Rho factor-dependent
 |
|---|
| © Daniel L.Kaplan, et al |
- The amino-terminal domain of Rho factor (blue) binds to the RNA sequence of Rho utilization site (rut). (Step 1)
- The carboxy-terminal domain of Rho (orange) binds the mRNA downstream from rut, and the Rho hexameric ring closes. (Step2)
- Ring closure propels Rho moving close to RNAP (Step 3).
- Rho dis-engages RNA and RNAP from DNA. (Step 4).
Eukaryotic Transcription
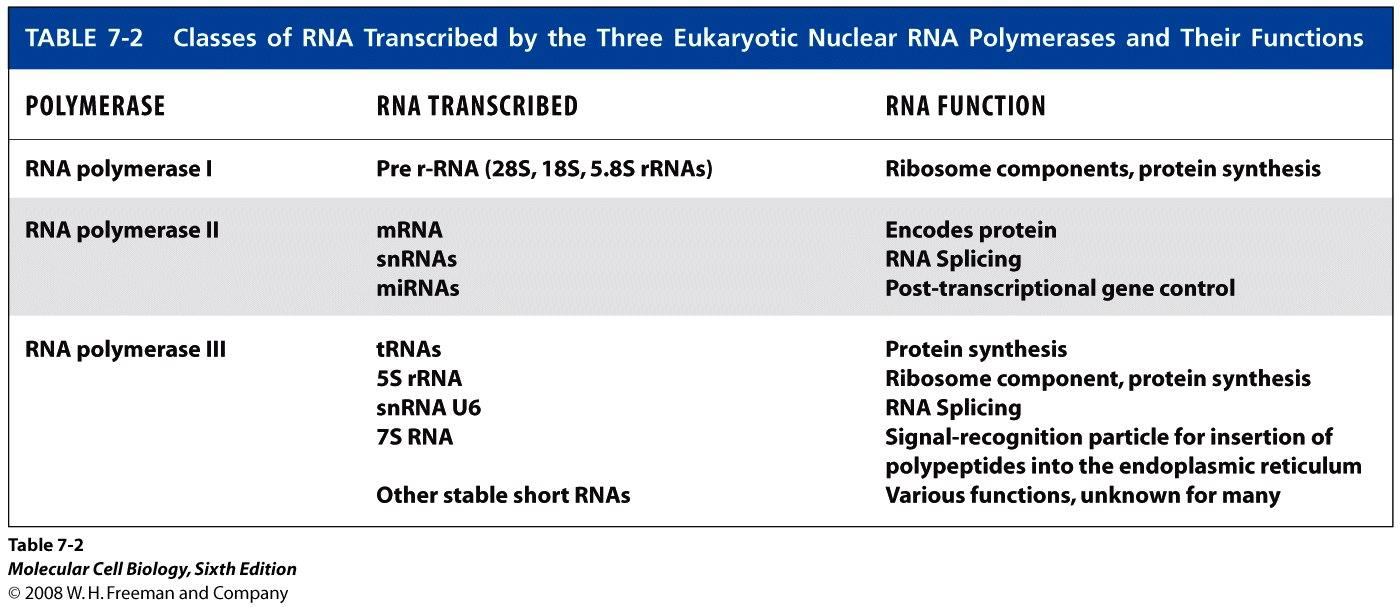
Three types of nuclear RNAPs
 |
|---|
Subunit compositions of nuclear RNAPs
- All three yeast RNAPs have five core subunits, similar to those of E. coli RNAP.
- α-like subunits different between RNAP-II and RNAP-I, -III
- Four other subunits are shared
Eukaryotic Transcription – RNAP II
 |
|---|
| © Karim-Jean Armache |
Yeast RNAP II structure resembling bacterial RNAPs
- A crab claw-like shape
- Positions and core folds of homologous subunits
- Negatively charged surface except for the DNA-binding cleft
Eukaryotic Transcription
RNA biosynthesis:
- The “clamp” of the Rpb1 subunit moves down to trap DNA between the two claw
- Unwind DNA at the active site
- The “wall” of Rpb2 kinks the template by 90° out of the active site
- One base of the template points at the active site (magnesium)
- This base is paired with the ribonucleotide base of RNA above a “pore” at the end of a “funnel” to the protein exterior (NTP entrance)
- A rudder loop of the clamp separates the RNA-DNA hybrid helix, allowing the dsDNA to reform.
RNA translocation:
- The “bridge” helix of Rpb1 contacts the DNA base at the active site
- The bridge helix switches between straight and bent conformation
- Bridge helix bending pushes away the DNA-RNA bases at the active site
- The straight conformation yield an empty pocket for entry of the new NTP
Transcription Inhibition as a Therapeutic Target for Cancer
- mRNAs of many oncogenes, as well as regulators of other key processes such as cell proliferation, angiogenesis, and apoptosis, typically have shorter half-lives.
- Transformed and non-transformed cells exhibit the differential sensitivity to RNA-directed agents.
Inhibiting transcription
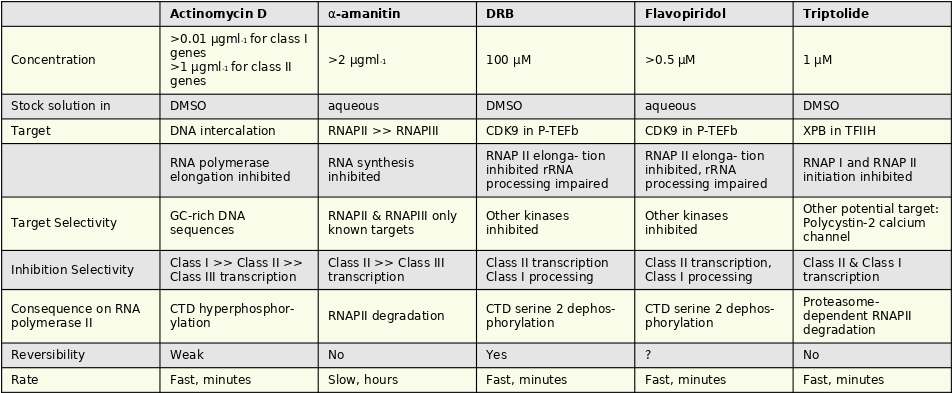 |
|---|
| Source Unknow |
α-amanitin
 |
 |
|---|---|
| © Srivindhya Kolluru, 2019 | © David A. Bushnell, 2002 |
Eukaryotic Transcription promoters
 |
|---|
| Source Unknow |
Mammalian Gene and Proximal Promoter (class II)
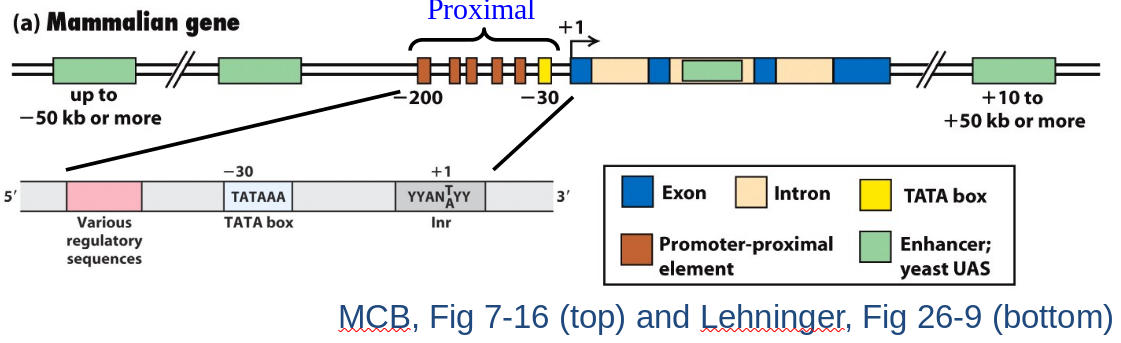 |
|---|
Proximal Promoter: Within 200 bp of transcription start.
- TATA Box (TATAAA)
- -30 upstream of start +1); fixed position.
- Recruits TATA Binding Protein (TBP) complex to promoter.
- Major assembly point for Transcription Preinitiation Complex (PIC).
- Initiator Region (Inr): Sequence where DNA is unwound. +1 usually within or near.
- Other activation elements:
- Critical for docking basal transcription apparatus.
- Tissue-specific elements.
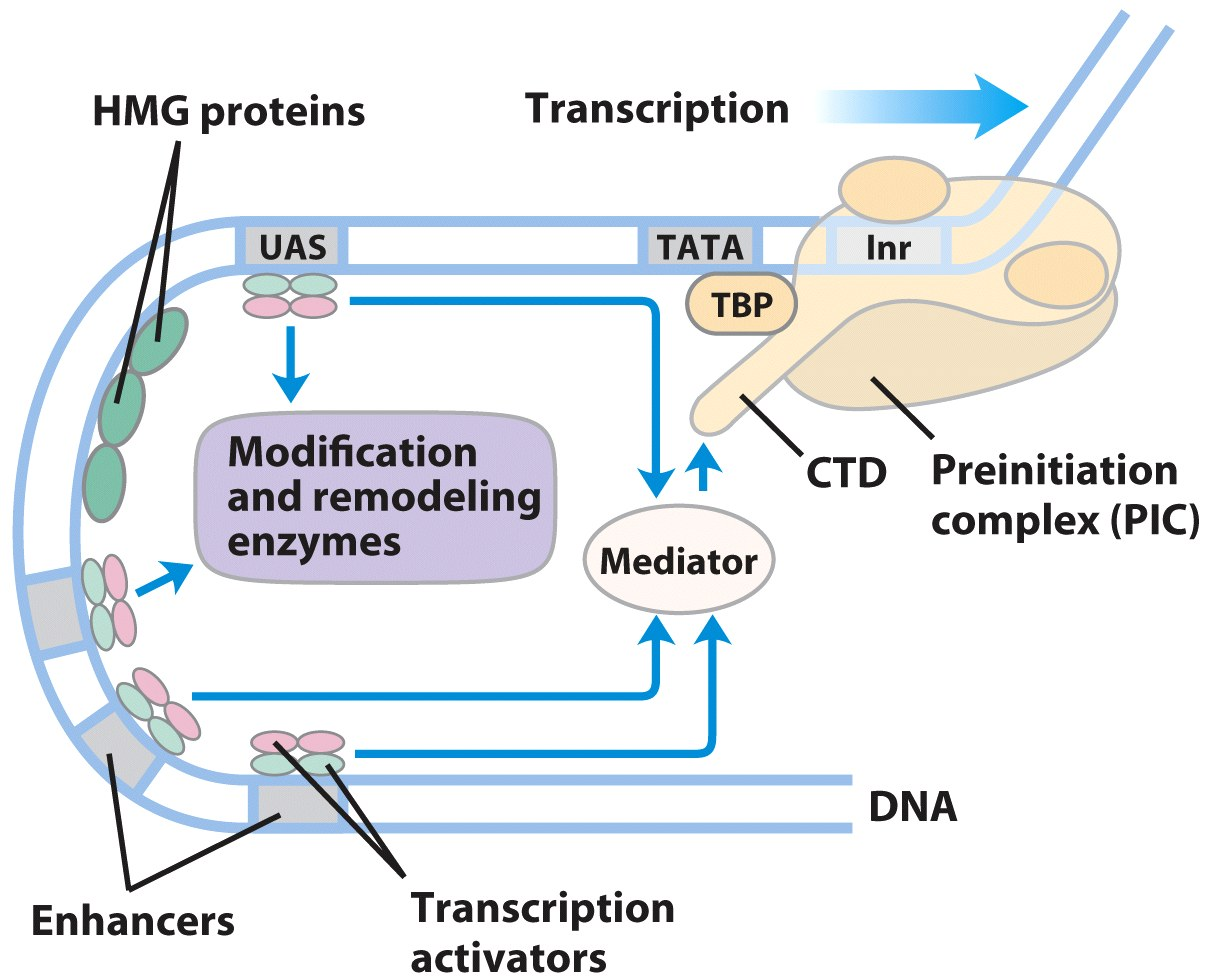
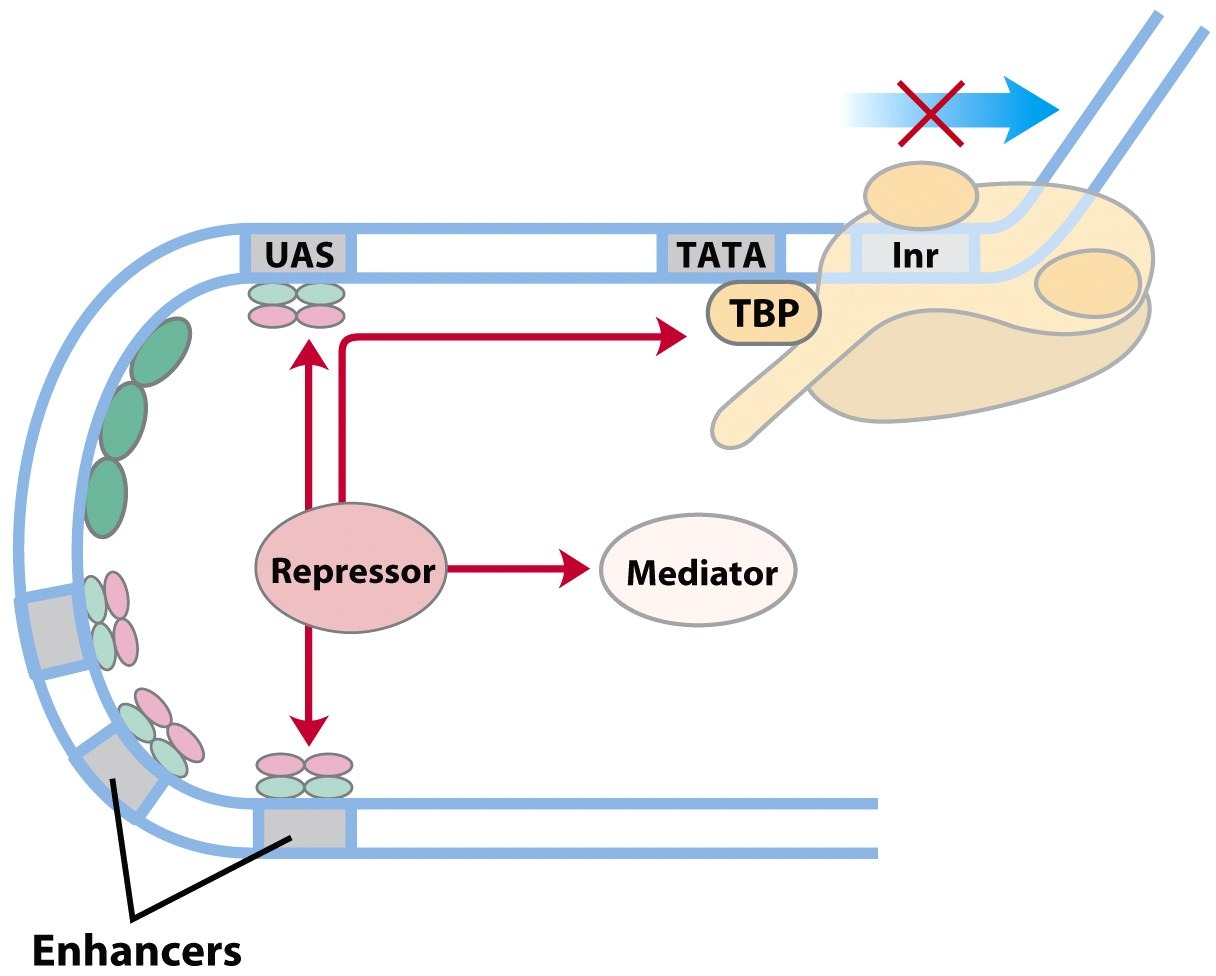
Eukaryotic Promoter with Activator Proteins & Repressor
| Activator Proteins | Transcriptional Repression |
|---|---|
 |
 |
| © Lehninger, Fig 28-29 | © Lehninger, Fig 28-29 |
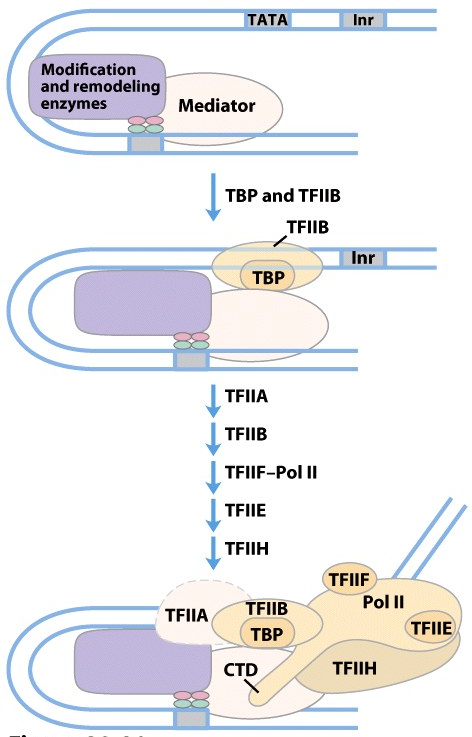
How is PIC Assembled?
 © Lehninger, Fig 28-30 |
Six transcription factors (TFs) for class II promoters
- Equivalent of a bacterial σ factor
- Highly conserved from yeast to human
- Required for the synthesis of all mRNAs
- Allow a low (basal) level of transcription, which can be increased by the gene-specific activators
Phospho-code on RNAP II CTD
 |
|---|
| © Sylvain Egloff, 2008 |
Non-canonical functions of transcription
- CENP-A defines centromeric chromatin
RNAP II transcription facilitates the deposition of CENP-A into centromeric chromatin
 |
|---|
| © F. Lyn Chan, 2012 |
- ChIP-Seq: determine protein distribution on chromatins
- RNA-Seq: determine the expression patterns of genes and other DNA sequences
Inhibitors of DNA synthesis and their specificities
AZT and DDI: reverse transcriptase, chain terminate
nuceoside analogues of guanosine: phosphorylated by a viral kinase
Polymerase processivity Enhanced by a Clamp
β clamp in bacteria
PCNA in eukayotes
loaded onto dsDNA on calmp
Clamp loader couples ATP Hydrolysis to Clamp Loading
Sliding clamp + ATP: open and bind DNA; ATP → ADP, DNA releases
Biological advantages of DNA
- Provides stable, yet mutable storage of genetic information. Metabolic stability and availability of repair mechanisms ensure long-term survival under a variety of physiological conditions.
- Serves as a template for accurate and adaptable replication of genomes. This biological role ensures the transmission of important physiological functions for multiple generations.
- Serves as a template for the expression and regulation of genetic information. This biological role facilitates adaptation of genomes to changing physiological environments.
- Amenable to diversification (i.e., can evolve). Mutation and genetic recombination (DNA rearrangements) are the two major driving forces in the evolution of genomic diversity and adaptation in nature.
DNA ligases
Eukaryotic DAN ligase use ATP rather than NADP
initiation od DNA replication in E. coli
origin region (OriC) ~245bp
Duplex unwinding by DNA helicase
- Exposed ssDNA allows SSB and helic …
Initiation: OriC-DnaA intercation → Entry of SSB and DnaB (hexlicase) → ENtry of promase → RNA primers are synthesized and the elongation complexes for 2 DNA replication forks are assembled
Elongation:
Prokaryotic DNA replication
A typical DNA replicsome
synthesis of leading strand are coupled with lagging strand by clamploader complex.
The e.coli Pol III holoenzyme …
Pols enyzme subunit factors
Eukaryotic DNA replication:
Pro: no cell cycle and start every time
Eu: onece anc only once in S phase; highly regulated
G1: Preparing
Checkpoint: end of the S phase
Time is variatd
Eukaryotic chromaomse have multiple origins…
Pro: single initial site
Euk: multiple initial site
Eukaryotic replication initiation is hihgly oreder… well regulated and complex…
Helicase (Mcm2-7)
Sld2/3 phosphorylated by CDK
Super Current Biochemical Reconstitution of Regulated replication origin firing
Termination in prokaryotes
Ter sites, 2 converging new ds circular DNA molecules separated from each other by recombination and with assistance of
Cell are constantly exposed to agents that can cause damage to their genome
Repair, tolerant, destroy the cell
Types of mutation
Transition: AT → G/C
Transversion: A/T → C/G
Frameshift: GGGGGG → GGGGG
Base alteration
Oxidation:
8-oxodG(antl)-dC(antf)
8-oxodG(synl)-dA(antf)
The rotation of the bound which cause the shift of the side chain and attribute difference hydrogen bonds. 3 to 2, C to A
Alkylation; Cross-links
Exogenous Damage: Exposure to ultraviolet radiation
Absorbed by adjacent pyrimidine bind and form double bond and cause mutation or lethal during replication/transcription
Repair
- Base Excision Repair
- Single Nucleotide
- Nucleotide Excision Repair
- oligonucleotide
- Mismatch Excision Repair
- mismatched base
Base Excision Repair
- how recognize
- AP site
- Donw string/ leading string doesn’t matter
- ligates involved
Nucleotide excission repair (NER)
- recognize Enzymes (XPC)
- recruit other factors
- unwinding, mutation is exposed
- Some protein was recruited and mistake was cut / rescued.
Recognition → resynthesis
Xeroderma pigmentosum (XP)
Accelerated skin cancer; mortality likely due to metastatic melanoma or squamous cell carcinoma
MMR pathway - E. Coli
MutS, MutL, MutH, ATP
MutH: cut the strand (down/leading)
methelation is critical here
RNA polymerases
-
Transcription using DNA to RAN
- Bacterial: ON enzyme for all of RNA synthesis
- Eukaryotic: At leased Three I II III
-
Strangs 1:
- Template;
- Non-coding;
- Anti-sense:
- Completed strand
-
Strands 2:
- Sense strand: coding strange of the DNA
- Non-completed strand
-
Bacterial promoter consensus sequences
- UP Element; -35; -10; RNA stra +1
- -35; -10: promoters (similar to regulatories in Eukaryotic.)
Expression controlled by interactions of promoter elements with RNA polymerase and specific repressor/Activators
Bacterial RNAP
- Holoenyme versis core enzyme
- Core subunits: α₂ββ’ω (400kDa)
- Holoenzyme: core subunits plus factors
- …
Five σ factirs in E. coli gene expression
- Exp: (not important)
- σ 70: Most genes
- σ 32: Heat schock proteins
- Trascription Elongation
- RANP trancks along the DNA template, synthesizes mRNA in te 5’ to 3’ direction and unwinds and rewinds the DNA as it reads.
- Transcription elongation causing DNA supercoiling, relaxed by topisomerases
- Termination
- Transcription Terminattion
- A series of 4 - 10 base
- Sequence depended: CGGCGCTTTTTT (CG rich region; AT rich region)
- ρ factor-dependent
- The amino-termina domain of ρ factor binds to hte RNA sequence of ρ utilization site
- the carboxy-terminal domain of ρ hexameric ring closes
- Ring closure propels ρ moving close to RNAP
- ρ dis-engages RNA and RNAP from DNA
- Transcription Terminattion
Eukaryotic Transcriotion
- Three types of Nuclears RNAPs
- I: rRNA (28S, 18S)
- II: mRNA; snRNAs; miRNAs
- III: tRNAs; 5S rRNA; snRNA U6 7S RNA; other stable short RNAs
- Transcription
- Subunit compositions of nueclear RNAPs
- β’; β; αI; αII; ω
- All yeast RNAP has five subunits similar to Bacteria
- RNAP II
- Yeast RNAP II strcture resembling bacterial RNAPs
- A vrab claw-like strcuture
- Yeast RNAP II strcture resembling bacterial RNAPs
- RNA biosynthesis
- The clamp of the Rpb1 subunit moves down to trap DAN between the two claw
- Unwind DNA at the active site
- Wall of Rpb2 kinks the template by 90° out of the active site
- One vase of the template points at the active site
- This base is paired with the ribonucleotide
- RNA translocation
- Transcription promoters
- Class I, II, II to RAN poly I, II, III
- Mammalian Gene and proximal promoter (Class II)
- Proximal region (-200 ~ -30), within 200 bp of transcription start; similar to the -10; -35 in the bacteria’s
- TATA Box (TATAAA)
- Recruits TATA binding protein …
- Initiator region (Inr)
- Other activation elements
- EPromoter and Activator proteins
- Transcriptional Activators: DNA binding domain; activation domain;
- Activator functions: Chromatin remodeling (acetylation-HAT); Mediator facilitates PIC assembly
- Repressor
- Bind UAS.enhancers → displace activators
- Prevent mediator → no PIC formation
- Attracts HDACs and HMTs → heterochromatin
- PIC Assmbled
- mediator facilitates TBP and TFIIB binding promoter
- Other basal TFs and Pol II bind
- phosphorylation of the CTD of Pol II by TFIIH → Transcription initiation
- Six transcription factors for clas II promoters (Not important)
- ==Phospho-code on RNAP II CTD
- TFIH: Phosphate the Ser5 would initiated the transcription activity
- P-TEFb: phospholate Ser2 to maintain the elongation state
- Non-canonical functions of transcription
- CENP-A defines centromeric chromatin
- RNA…
- ChiP-Seq: Determine protein distributed on chromatins
- RNA-Seq: Expression profiles
- Subunit compositions of nueclear RNAPs
Transcription inhibition as a therapeutic target for cancer
- mRANs of many oncogenes
- Transformed → RNA-directed agents: different sensitivity.
- Alpha-amanitin: from mushroom
- directed binding to RNAP, adn block the bridge to repress the conformation change
Bacterial Transcription
Eukaryotic transcription
RNAP II CTD phospho-code
Non-canonical functions of transcription
Assays to study transcription
posttranscriptional Modification of Eukaryotic RNAs
- Role of 5’ cap
- Ribosomal recogintion during translation
- Cap structure
- 7-methylguanosin (m⁷G)
- joined to the mRNA first nucleotide
- Via a 5’-5’ tri-P brifge
- Involved Enzymes
- RNA riphosphatase removing the γ-P of the mRNA’s 5’ site
- mRNA guanlyltransferase (a capping enzyme) adding GMP. (Take GMP to the site)
- Guanine-7-methyltranserase (second capping enzyme) methylating guanine
- Capping enzymes bind to RNAP-II, which will switch RAN synthesis initiation to elongation ()
posttransicriptional Modification of Eukaryotic RANs
- Poly A tails; ~250 nt
- Cleavage and polyadenylation specificity factor (CPASF), cleaving up to 35 nt past the AAUAAA sequence
- Poly A polymerase generate poly A tail using ATP. (termination signals given to RNA-polymerase)
- CPSF binds to RNAP-II, coupling polyadenylation to transcription termination.
- Poly A tail binds to poly-binding protein, protecting from degradation, increasing mRNA stability.
Exons and Introns
-
Precurso mRANs (pre-mRNAs) are processsed by the excision of introns and the splicing (joing) of exons.
-
Exon splicing in Two-Stage Reactions
- Invariant Sequences for splice junction
- GU at the intron’s 5’
- AG at the intron’s 3
- (graph), the number under the NT means the ration you are supposed to see them on the Intron
- A branch point near the 3’ splice site
- Free G, not paire to the intron…
- A2’-5’ P-diester bind between the tinron A (OH²’) and the intron at the 5’ splice site, forming a “lariat” structure.
- The Exon 1 OH³’ group at the 5’ splice site from a 3’-5’ P-diester bind with the Exon 2’ at the 3’ splice site, releasing the intron with the free OH³’ group.
- The intron keeps the lariat structure.
- Note: Splicing proceeds w/o free energy lose, Cleavage of one P-diseter bond and formation of a new bond.
- Invariant Sequences for splice junction
Spliceosome-aided RNA splicing
Splicing could be focilited by the protein Splicesome.60S spliceosome particle containing five small nuclear RNAs + Ribonucleoproteins (U 1~6, no 3)
- U1 recognizes the 5’ splice junction
- U2 recognizes the branch point (intron A)
- The binding of U4-U5-U6 forms spliceosome.
- RAN cleavage at the 5’ splice site
- RNA cleavage at the 3’ splice site
- Intron …
Self-Splicing RNA
- Group II Intron
- Exson 1 and Exon II aligned together
- the giant rondant was splicesd by themselves
- Group I intron:
- not Intron A to initiated the reaction
- G , GTP, GDP, GMP… as the starting site
- The 3’-OH of …
Summary of spliceosome-aided Splicing and Self-Splicing
- In goup I introns, aguanosine cofactor (G) tat is not part of the RNA chain associates with the active site. The 3’-hydroxyl group of this G attacks the 5’ splice sit;
- The reaction is similar to those involving the 2 hydroxyl groups of branch sites as in group II introns and RNA inrons spliced in spliceosomes.
- The subsequent trans-e…
Interactions of RNAP-II with capping enzyme, splicing and CPSF
Figure 16-5
Aminoacyl-tRAN Synthetase
- Attach amino acids to tRNAs
- Two step reactions
- Amino acid activation by the reaction with ATP
(Aminoacyl-tRNA syntheiase) - Formation of an aminoacyl-tRNA
- Amino acid activation by the reaction with ATP
Aminoacyl-tRAN SYnthetases
- The synthetase enzymes have an elongated shape.
- Binding the anticodon of tRNA near the one end of the enzyme.
- Binding the A.a. acceptor stem of tRNA near th other end
Proofreading by Aminoacyl-tRAN Synthetase
Binding to the amino acid substrate pocket of tRNA Synthetase that consists of the zinc ion.L-Group: Megan, Ryan, Ka
-
Threonine binds at this pocket b/c it can coordinate Zn2+ using it NH2 and OH groups
-
tRNAthr is tRNA for threonin.
- Attaching ThR to tRNAthr makes Thr-tRNAthr (tRNA is correctly charged)
- Attaching Ser to tRNAthr makes Ser-tRNAthr (tRAN is mischarged)
Acetylation
- definition
- histone modification
- Acetylation by HATs
- Recognition by bromodomains
- Deacetylation by lysine deacytelases (HDACs adn Sirtuins)
- cofactors: Zinx, NAD
- Methylation by histone methyltransefrases (HMTs)
- The donor: S-adenosylmethionine (SAM)
- SET domains in SAM
- Recognition by chromodomains and plant homeodomain (PHD) fingers
- Aromatci cage: to recognize
- Chroodomain-containing heterochromatin protein 1 (HP1) function: regulated transcription
- Demethylation
- Lysine-specific histone demethylases (LSD1 and LSD2)-FAD-dependent
- Jumonji C-domain (JMJC) family members - F2a and α-ketoglutarate-dependent
Chromatin structure
Chrmosome → chromatin fiber → Beads on a string DNA wound on nucleosomes → Histones + Doble helix
- Heteochromatin: Highly condensed nonexpressing DNA. (PS: Canot be transcripted because transcript machinary can’t recognize and bind)
- Euchromatin: Less condensed, Transcriptionally active DAN
H2A, H2B, H3, H4
Histone Modification
Histone was circled in the center of the DNA, but with tail out of the stracture and cann’t be visualized by cristal structure. But they are the key site for the Methylating or Acetylating
acKL acetyl lysine
meR:
meK:
PS: phosphoryl serine
…
Histone Code
Histone modification is the signals of transcription on/off.
Histone Acetyltraseferases (HATs)
Histone + Me-CoA → CoA + Me-Histone
Structure of the HAT domain from Tetrahymena thermophila
- The enzyme is deeply clefted
- The histone H3 peptide KSTGGK14APRK! and coenzyme A are bound at the deep cleft.
Catalysis appears to involve water-mediated proton extraction from the substrate lysine by a glutamic acid general base.
Acetylated Lys recognition by Bromodomains
-
Bromodomains specifically bind acetylated lysine residues on histones.
- A deep hydrophobic pocket (hole) accommodates the acetyl-Lys side chain (PS, the hole recognize the acetyl-Lys specificity)
-
TAF1: has two brpmodomains and dipart from each with ~25Å, separated by 7 ~8 residues.
-
A subunit of transcription factor TFIID (PDBid=1eqf)
-
…
-
…
Role of TAF1 double Bromodomain
- TAF1 double bromodomain targets TFIID to promoters
- A refined proposal for transcription initiation
- A AHT-cotaining coactivator binds to upstream DAN binding protein (An activator)
- The HAT acetylates nearby histone tails
- TFIID is recruited to the site via the binding of the TAF1 double bromodomain to the acetyl-Lys residues of histones.
- followed by recruitment of other initiation factors and RNAP-II for transcription initiation.
Histone Deacetylases (HDACs)
- 18 HDAC enzymes that deacetylate acetyl lysine substrates including histones.
- Histone deacetylase …
- …
Reaction mechanism:
- Bound Zn2+ mediates the nucleophilic attack of water on acetylated lysine, forming a tetrahedral oxyanion intermidiate
- The carbon-…
Situins
NAD⁺-dependent reaction
- nucleophilic addition to form a C1’-O-alkylamidate intermediate and free nicotinamide.
- Teh 2’hydroxy group of the NAD⁺ ribose attacks the C1’-O-alkylamidate to form the 1’,2’cyclic intermediate
- The formation of deacetylasted lysine and 2’-O-acetyl-ADP ribose.
Thus, nicotinamide, a deacetylated lysine-containing histone, and 2’-O-acetyl-ADP
Histone Methylation
- Methylation at Lys and Arg of H3 and H4 tends to silence the genes, inducing heterochromatin formation
- However, trimetheylated Lys4 of H3 is associated with active genes
(methylation can add one to 3 methyl in a time) (give a exampel)
- Histone methyltransferases (HMTs) use S-adenosylmethionine (SAM) as a methy donor
- The HMT enzymes have a SET domain containing the catalytic site for methylation.
Structure of SET7/9 (PDB=1o9s)
SET7/9 mono-methylates Lys4 of H3
- SAM (SAH) adn the Lys4-containing peptide bind to oppsite sites of the protein
- The lys4 side chain is inserted through a narrow channel and positioned for methylation by SAM.
Methylated Histone Recognition
- Chromodomains and plant homeodomain (PHD) fingers bind to mono, di- or tri-methylated Lys by an aromatic cage.
- Fits in the shape,
- lots of electron to nutralize the positive charge of the methyl group???
Consequence of Methylated hisone Recognition
- The chromodomain heterochromatin protein 1 (HP1) bidns to methylated H3 Ly9, contributing to gene silencing,
- Bound HP1 recruitsthe HMT Suv39h, methylating nearby histone lysines (H3K9), thereby recruiting more HP1 and forming HP1 complexes.
- This mechanism explains how heterochromatin spread to silence neighboring genes.
- HP1 clust prefer to form a tide complex and so, silence the DNA transcription.
Histone Demethylases
LSD1 & LSD2 demethylases
- Flacin adenine dinucleotide (FAD)- dependent amine oxidation
- Demethylate mono- and dimethylated substrates
The JMJC family
- Dioxygenases dependent on Fe2+ and α-ketoglutarate
- Demethylates mono-, di- and trimethylated substrates
Focuse on the Chemical reaction
Understanding the demethylation reaction from the LSD1 Structure
The LSD1 is bound to a 21 amino acid H2 peptide with a mutation of Lys4 to Met.
Modeling predicts that methylated Lys3 binds in a solvent inaccessible area in front of the FAD cofactor (electron extraction).
Understanding … JMJD3 structure
THe JMJD3 is bound to a H3 pep with a trimetrhylated Lys27
- The cofactor analog N-oxylayglycine NOG not α-ketoglutarate and nickel not iron are used to prevent the reaction
The methylated Lys27 side chain insertes deep into the catalytic pocked, close to the cofactor analog and nickel for demethylation reaction.
Chromatin Writers, Readers and Erases
brif adn = =
Recombination
- homologous reconbination, need small similar sequnces
- Site-specific reconbination, nees specific DNA sequence
- ranspositions, no Specific DNA
- Non-homologous DNA end-joining (), specific proteins that repair dsDNA breaks
Homologous reconbination
- Required for accurate chomosome segregation
- repairing and tolerating DNA damage
- Recovery of stalled or broken replication forks
- Defects in recombination can contribute to genome instability and cancer predispositions
Biological roles
- Genetic reassortments in gametogenesis
- Repair of DNA damages
- Repair replication forks
- laterl DNA transfer between ceels
- therOther, deletions, inversions, translocations, etc.
How HR produces deletions, insertions and inversions
- Incersion:
DNA fold in a revised direction, The DNA form a arch bridge and at the foot of the bridge connected - Deletion and insertion
DNA fold in a same direction. A circle formed and the cross point was connected together and the circle was deleted. (Insertion happens the similar way.)
Translocation that casue cancers
- Chronic myeloid leukemia
BCR/ABL mRNA
p210 fusion protein
ABL are concertive hyregulated RTK, BRC is a higly active promoter which drive the high expression of the ABL which resbonsible to growth and differentiation.
Holliday Model of Recombination
Nicking → Strand Exchange → Branch Migration → Resoltion → Recombination molecules
SOS response: Mutagenic Survival Pathway of Last Resort
- DNA Damges → ssDNA or dsDNA break → No Transcription
Normal LaxA turning of LexA-Regulated gene - Auto-cleavage of LaxA recruitment lots of RecA.
RacA: Pre-synapsis → Synapsis → ppost-synapsis
RacA complex driving homology search and alignment of complementary sequences.
Site-specific reconbination: intergration of lambda phage DNA into the E.coli Chromosome
DNA Transcription, Transcription Factor Binding
https://karobben.github.io/2021/11/27/LearnNotes/tulane-biochem-15/








