IMGT®: the international ImMunoGeneTics information system®
What is IMGT
IMGT® provides a range of databases, tools, and web resources focused on the immune system, particularly on the genetic and structural aspects of immunoglobulins (IG), T cell receptors (TCR), major histocompatibility complex (MHC) of all vertebrate species, and related proteins of the immune system (RPI) of any species. These resources are crucial for research in various fields, including immunology, genetics, bioinformatics, drug design, and personalized medicine.
Key features and offerings of IMGT® include:
-
Databases: IMGT® offers several databases containing detailed information on IG, TCR, and MHC sequences and structures, along with RPI. These databases are meticulously curated and regularly updated.
-
Analysis Tools: IMGT® provides tools for sequence analysis, gene identification, and 3D structure determination. IMGT/V-QUEST, for instance, is a widely used tool for the analysis of IG and TCR sequences.
-
Standardized Nomenclature: IMGT® has established a standardized nomenclature for the description of IG and TCR genetic components, which is essential for consistent communication and research in the field.
-
Educational Resources: The system also offers educational resources for those new to the field of immunogenetics, including tutorials, glossaries, and comprehensive descriptions of the molecular components of the immune system.
-
Research and Clinical Applications: The information and tools provided by IMGT® are invaluable for various applications, including research in immunology, genetics, and autoimmunity, as well as in clinical settings for antibody engineering, diagnosis, and understanding of immune disorders.
Main Service of the IMGT
| Category | Name | Description/Focus Area |
|---|---|---|
| Databases | IMGT/GENE-DB | Database for immunoglobulin (IG) and T cell receptor (TCR) genes of all vertebrate species. |
| IMGT/3Dstructure-DB | Database for 3D structures of IG, TCR, MHC, and RPI (related proteins of the immune system). | |
| IMGT/LIGM-DB | A comprehensive database of IG and TCR nucleotide sequences from various species. | |
| IMGT/PRIMER-DB | Database of primers and probes for IG and TCR gene sequences. | |
| IMGT/PROTEIN-DB | Database for IG, TCR, MHC, and RPI protein sequences and structures. | |
| Analysis Tools | IMGT/V-QUEST | Tool for the analysis of IG and TCR nucleotide sequences. Identifies V, D, and J gene segments and alleles. |
| IMGT/JunctionAnalysis | Tool focused on detailed analysis of the V-J and V-D-J junctions of IG and TCR sequences. | |
| IMGT/HighV-QUEST | High-throughput version of IMGT/V-QUEST for next-generation sequencing (NGS) data. | |
| IMGT/DomainGapAlign | Tool for the analysis of IG, TCR, and RPI domain sequences and comparison with IMGT reference directory. | |
| IMGT/Collier-de-Perles | Tool for two-dimensional (2D) graphical representation of IG and TCR variable domains. | |
| Resources | IMGT Education | Educational resources, including tutorials, glossaries, and comprehensive descriptions of immunogenetics. |
| IMGT Scientific chart | Standardized nomenclature and classification for IG, TCR, and MHC of humans and other vertebrates. | |
| IMGT Repertoire | Compilation of allelic polymorphisms and protein displays for variable (V), diversity (D), and joining (J) genes. | |
| Other Services | IMGT/Therapeutic | Information on therapeutic antibodies and fusion proteins for immune applications. |
| IMGT/mAb-DB | Specific database for monoclonal antibodies (mAbs). |
What is IMGT/V-QUEST database?
IMGT/V-QUEST is a specialized database and analysis tool that is part of the broader IMGT®, the international ImMunoGeneTics information system®. This system is a high-quality integrated knowledge resource specializing in immunoglobulins (IG), T cell receptors (TCR), major histocompatibility complex (MHC) of all vertebrate species, and related proteins of the immune system (RPI) of any species.
IMGT/V-QUEST specifically provides detailed analysis of nucleotide sequences for immunoglobulins (IG) and T cell receptors (TCR). It is widely used in immunology and related research fields for tasks such as:
-
Sequence Analysis: It allows for the identification and delimitation of V, D, and J genes and alleles in the input sequences. This is crucial for understanding the genetic basis of the immune response.
-
Clonotype Characterization: Researchers use it to characterize the clonotypes (unique T cell or B cell receptor sequences) in an individual’s immune repertoire, which is important in studies of immune system diversity and response.
-
Somatic Hypermutation Studies: It helps in the analysis of somatic hypermutations, which are critical for understanding adaptive immunity and processes like affinity maturation.
-
Comparative Immunology: By providing a comprehensive and curated database of IG and TCR sequences across different species, it aids in comparative immunology studies.
As of my last update in April 2023, IMGT/V-QUEST continues to be a valuable resource for immunologists, molecular biologists, and other researchers studying the adaptive immune system. The database is regularly updated to include new findings and sequences, ensuring its relevance and usefulness in the field.
Something You May Want to Know
c16 > g, Q6 > E (++−) means that the nt mutation (c > g) leads to an AA change at codon 6 with the same hydropathy (+) and volume (+) but with different physicochemical properties (−) classes ( 12 )[^Pommié_04].
What is IMGT-Ontology?
IMGT/Ontology is a key component of IMGT®, the international ImMunoGeneTics information system®. It represents the first ontology for immunogenetics and immunoinformatics and is a foundational aspect of the IMGT® information system. Developed by Marie-Paule Lefranc and her team, IMGT/Ontology provides a standardized vocabulary and a set of concepts that are essential for the consistent annotation, description, and comparison of the immune system’s components across different species. So, IMGT/Ontology is more of a set of criteria or a framework rather than a specific tool or software.
Key features and aspects of IMGT/Ontology include:
1. Standardized Vocabulary
IMGT/Ontology provides a controlled vocabulary for the description of immunogenetic sequences. This includes terms for genes, alleles, and other genetic elements relevant to immunogenetics.2. Classification Criteria
It establishes clear and standardized criteria for the classification of immunoglobulins (IG), T cell receptors (TCR), and major histocompatibility complex (MHC) molecules. This classification is crucial for the accurate comparison and analysis of these molecules.3. Identification of Key Concepts
The ontology identifies and defines key concepts in immunogenetics, such as gene segment functionality, recombination features, and junctional diversity. These concepts are critical for understanding the adaptive immune response.4. IMGT-ONTOLOGY Axioms
These are the rules that define the relations between the concepts and are used for the annotation and description of the IG, TCR, and MHC sequences in the IMGT® databases and tools.5. Facilitating Data Integration and Analysis
By providing a common framework and language, IMGT/Ontology enables the integration, analysis, and sharing of immunogenetic data across different research projects and databases.6. Support for Research and Clinical Applications
The standardized approach of IMGT/Ontology supports various research and clinical applications, including antibody engineering, vaccine development, and the study of immune responses.IMGT-ONTOLOGY axioms and concepts
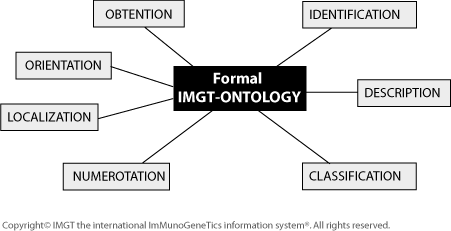
Seven IMGT-ONTOLOGY axioms have been defined: ‘IDENTIFICATION’ [1], ‘DESCRIPTION’ [2], ‘CLASSIFICATION’ [3], ‘NUMEROTATION’ [4][5], ‘LOCALIZATION’, ‘ORIENTATION’, and ‘OBTENTION’. They constitute the Formal IMGT-ONTOLOGY or IMGT-Kaleidoscope [6].
 |
|---|
| © IMGT Education |
IMGT/Ontology is a critical resource for researchers and professionals in immunology, genetics, bioinformatics, and related fields. It ensures that data and analyses are consistent, reproducible, and interoperable, which is vital in advancing our understanding of the immune system and in developing immunotherapy and other medical applications.
The oldest paper about ontology:
- 1999: Ontology for immunogenetics: the IMGT-ONTOLOGY [7]
- The first ontology to be developed for immunogenetics. ‘An ontology is a specification of conceptualization’ (Gruber, 1993).
- Covers four main concepts: ‘IDENTIFICATION’, ‘DESCRIPTION’, ‘CLASSIFICATION’ and ‘OBTENTION’.
- 2008: IMGT-Kaleidoscope, the formal IMGT-ONTOLOGY paradigm[8]
- seven axioms, ‘IDENTIFICATION’, ‘DESCRIPTION’, ‘CLASSIFICATION’, ‘NUMEROTATION’, ‘LOCALIZATION’, ‘ORIENTATION’ and ‘OBTENTION’
References
- IMGT®: Ontology (IMGT-ONTOLOGY)
- Duroux P, Kaas Q, Brochet X, et al. IMGT-Kaleidoscope, the formal IMGT-ONTOLOGY paradigm[J]. Biochimie, 2008, 90(4): 570-583.
- Giudicelli V, Lefranc M P. Imgt-ontology 2012[J]. Frontiers in genetics, 2012, 3: 79.
- Lefranc M P. Immunoglobulin and T cell receptor genes: IMGT® and the birth and rise of immunoinformatics[J]. Frontiers in immunology, 2014, 5: 22.
Other References
Lefranc, M.-P. From IMGT-ONTOLOGY IDENTIFICATION Axiom to IMGT Standardized Keywords: For Immunoglobulins (IG), T Cell Receptors (TR), and Conventional Genes. Cold Spring Harb Protoc., 1;2011(6): 604-613. pii: pdb.ip82. doi: 10.1101/pdb.ip82(2011) PMID:21632792. ↩︎
Lefranc, M.-P. From IMGT-ONTOLOGY DESCRIPTION Axiom to IMGT Standardized Labels: For Immunoglobulin (IG) and T Cell Receptor (TR) Sequences and Structures. Cold Spring Harb Protoc., 1;2011(6): 614-626. pii: pdb.ip83. doi: 10.1101/pdb.ip83 (2011) PMID:21632791. ↩︎
Lefranc, M.-P. From IMGT-ONTOLOGY CLASSIFICATION Axiom to IMGT Standardized Gene and Allele Nomenclature: For Immunoglobulins (IG) and T Cell Receptors (TR). Cold Spring Harb Protoc., 1;2011(6): 627-632. pii: pdb.ip84. doi: 10.1101/pdb.ip84 (2011) PMID:21632790. ↩︎
Lefranc, M.-P. “IMGT Collier de Perles for the Variable (V), Constant ©, and Groove (G) Domains of IG, TR, MH, IgSF, and MhSF” Cold Spring Harb Protoc. 2011 Jun 1;2011(6). pii: pdb.ip86. doi: 10.1101/pdb.ip86. PMID: 21632788 ↩︎
Lefranc, M.-P. IMGT Unique Numbering for the Variable (V), Constant ©, and Groove (G) Domains of IG, TR, MH, IgSF, and MhSF. Cold Spring Harb Protoc., 1;2011(6). pii: pdb.ip85. doi: 10.1101/pdb.ip85 (2011) PMID: 21632789 ↩︎
Duroux P et al., IMGT-Kaleidoscope, the Formal IMGT-ONTOLOGY paradigm. Biochimie, 90:570-83. Epub 2007 Sep 11 (2008) PMID:17949886 ↩︎
Giudicelli V, Lefranc M P. Ontology for immunogenetics: the IMGT-ONTOLOGY[J]. Bioinformatics, 1999, 15(12): 1047-1054. ↩︎
Duroux P, Kaas Q, Brochet X, et al. IMGT-Kaleidoscope, the formal IMGT-ONTOLOGY paradigm[J]. Biochimie, 2008, 90(4): 570-583. ↩︎
IMGT®: the international ImMunoGeneTics information system®