Structure of the Immunoglobulin
The Structure of the Immunoglobulin
 |
|---|
| © Adrian Y. S. Lee |
Immunoglobulins, commonly known as antibodies, are crucial proteins in the immune system that recognize and bind to specific antigens, such as bacteria and viruses, to help protect the body. Their structure is both unique and complex, consisting of several key components:
-
Basic Structure: Immunoglobulins are Y-shaped molecules made up of four polypeptide chains - two identical heavy (H) chains and two identical light (L) chains. These chains are held together by disulfide bonds.
-
Variable (V) and Constant © Regions:
- Variable Regions: The tips of the ‘Y’ shape consist of the variable regions of the light and heavy chains. These regions are highly diverse and are responsible for the antigen-binding specificity of the antibody.
- Constant Regions: The rest of the molecule forms the constant region, which is relatively conserved across different antibodies. The constant region of the heavy chains determines the class or isotype (e.g., IgG, IgM, IgA, IgE, IgD) of the antibody and mediates effector functions.
-
Isotypes: Mammals have several classes of immunoglobulins (IgG, IgA, IgM, IgE, IgD), each with different roles in the immune response. These isotypes differ mainly in their heavy chain constant regions.
-
Glycosylation: Many antibodies are glycosylated, meaning they have carbohydrate groups attached. This glycosylation can affect the antibody’s stability, distribution, and activity.
-
Light Chain Types: There are two types of light chains in antibodies - kappa (κ) and lambda (λ). An individual antibody will have two identical light chains of one type.
V(D)J
 |
|---|
| © David B. Roth |
V(D)J recombination is a mechanism in the immune system that generates the immense diversity of antibodies (immunoglobulins) and T cell receptors necessary for the adaptive immune response. This process is named for the three gene segments involved in the recombination: Variable (V), Diversity (D), and Joining (J). So, V(D)J is the recombination unit.
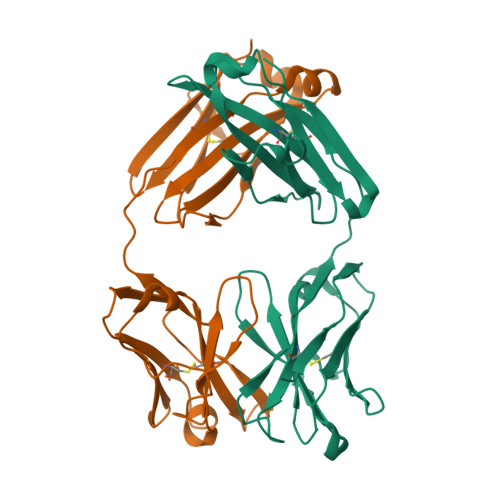
V(D)J in 3D Structure
We take 5wl2 as example:
 |
|---|
| ©PDB 5wl2 |
Kabat Number
 |
|---|
| © pipebio |
The Kabat definition is based on sequence variability and is the most commonly used. Here is the CDR definition from Bioinf.org.uk
| L1 | L2 | L3 | H1 | H2 | H3 | |
|---|---|---|---|---|---|---|
| Kabat | L24-L34 | L50-L56 | L89-L97 | H31-H35B | H50-H65 | H95-H102 |
How to make it in python:
|
|
Structure of the Immunoglobulin
https://karobben.github.io/2024/01/02/LearnNotes/igstructure/








