Antibody 12/23 rule
How Does Antibody Fragments Jointed Together?
We all know that antibodies are composed of V, D, and J segments, which originate from different locations on the chromosome. But how are they connected together after post-transcriptional modification? The 12/23 rule is the fundamental mechanism that ensures proper recombination of these segments.
The V region, or V domain, of an immunoglobulin heavy or light chain is encoded by more than one gene segment. For the light chain, the V domain is encoded by two separate DNA segments. The first segment encodes the first 95–101 amino acids of the light chain and is termed a V gene segment because it encodes most of the V domain. The second segment encodes the remainder of the V domain (up to 13 amino acids) and is termed a joining or J gene segment[1].
What Is 12/23 Rule
Video tutorial: Daniel Levy; 2013. VDJ Gene Recombination
The 12/23 rule is a principle in V(D)J recombination, a process crucial for generating the diversity of antibodies and T-cell receptors. It states that recombination can only occur between gene segments flanked by recombination signal sequences (RSS) with spacers of 12 base pairs (bp) and 23 bp. This ensures proper alignment and prevents inappropriate recombination events, maintaining the integrity and functionality of the immune system’s response.
 |
|---|
| © Charles A. Janeway |
This image illustrates the 12/23 rule in the context of V(D)J recombination.
-
Recombination Signal Sequences (RSS):
- Heptamer: A conserved sequence of 7 base pairs.
- Nonamer: A conserved sequence of 9 base pairs.
- Spacer: The region between the heptamer and nonamer, either 12 or 23 base pairs long.
-
V(D)J Recombination Process:
- Segments: V (Variable), D (Diversity), and J (Joining) segments.
- Rule Application: The 12/23 rule ensures that only segments with RSS of different spacers (12 and 23 bp) recombine, facilitating the correct assembly of these segments.
The image visually represents how the rule guides the alignment and recombination of V, D, and J gene segments, which is essential for generating the diversity of antibodies and T-cell receptors.
How It Worked
The heptamer and nonamer is the recombination signal sequences (RSSs). The (RAG1-RAG2)2 endonuclease complex (RAG) specifically recognizes and cleaves a pair of RSSs[2]
 |
|---|
| © Ru H[2:1] |
How Does It Applied
IgDetective
IgDetective, published by Vikram Sirupurapu and Yana Safonova[3], is a tool for detecting and naming antibodies based on the 12/23 rule. This tool leverages the stringent application of the 12/23 rule among mammals, making it suitable primarily for mammalian species.
- Test: not suitable for birds like chicken
williamdlees-Digger
This is another open source tool developed by William D. Lees[4], aimed at annotating the positions of V/D/J genes on newly assembled genomes. According to the results shown in the documentation, it has very high accuracy.
Prerequisite:
- a set of known core coding region allele sequences
- Position-weighted matrices
Some technic details worthy to know:
- The search for V-sequences uses the parameters gapopen 5, gapextend 5, penalty -1, word_size 11.
- D and J searches, word_size is reduced to 7 to reflect the shorter sequences
- D sequences the evalue is set to 100 rather than the default of 10 to widen the search.
How it find the heptamers and nonamers:
| Type | Functionality criteria |
|---|---|
| V | RSS nonamer and heptamer pass PWM thresholds, and match canonical consensus, if defined. L-PART1 and L-PART2 pass PWM thresholds and splice to form a coding sequence with no STOP-CODONs that is in-frame with the V sequence. V-REGION is in-frame and has first and second cysteines at the correct positions in the IMGT alignment. No STOP-CODONs are present in the V-REGION before the second cysteine. |
| D | RSS nonamers and heptamers match canonical consensus, if defined. |
| J | RSS nonamer and heptamer pass PWM thresholds, and match canonical consensus, if defined. The J-motif is found at the expected position relative to the end of the J sequence. The donor splice is found at the expected position, given the length of the matched sequence. |
Limitation: It only supports IMGT well-annotated species because it relies on germline annotation from the IMGT database.
Merits: Easy used and to be understood (write by python).
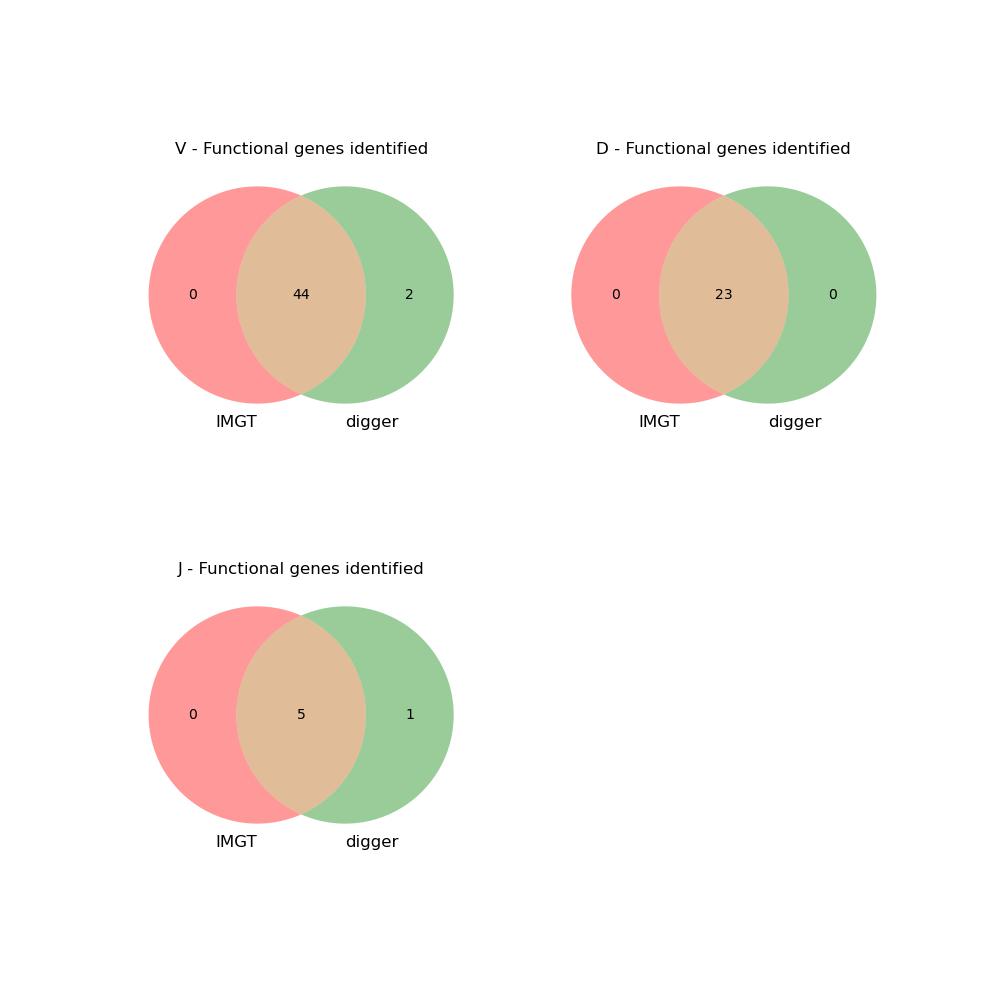 |
|---|
| © William D. Leeszs |
|
How It Really Looks Like in Human Genome
I randomly checked a few sequences from the homo genome and found that those regions from different V, D, and J genes are very similar. It could because that the 12/23 rule is not very stringent but flexible. But it could also caused by they are prevented to be recombined.

Janeway C, Travers P, Walport M, et al. Immunobiology: the immune system in health and disease[M]. New York: Garland Pub., 2001. ↩︎
Ru H, Zhang P, Wu H. Structural gymnastics of RAG-mediated DNA cleavage in V (D) J recombination[J]. Current opinion in structural biology, 2018, 53: 178-186. ↩︎ ↩︎
Sirupurapu V, Safonova Y, Pevzner P A. Gene prediction in the immunoglobulin loci[J]. Genome research, 2022, 32(6): 1152-1169. ↩︎
Lees W D, Saha S, Yaari G, et al. Digger: directed annotation of immunoglobulin and T cell receptor V, D, and J gene sequences and assemblies[J]. Bioinformatics, 2024, 40(3): btae144. ↩︎
Antibody 12/23 rule
https://karobben.github.io/2024/05/18/LearnNotes/ab1223rule/








