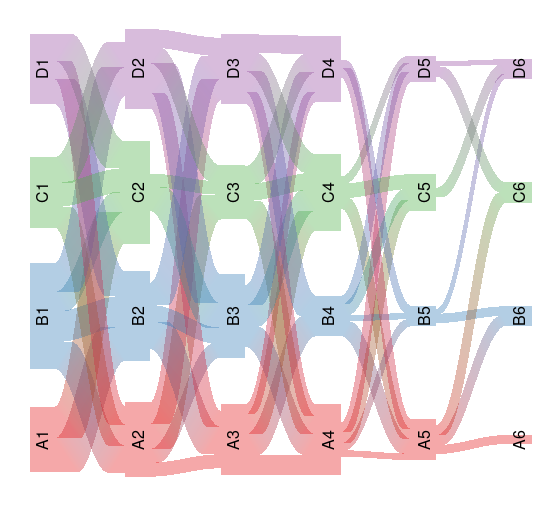
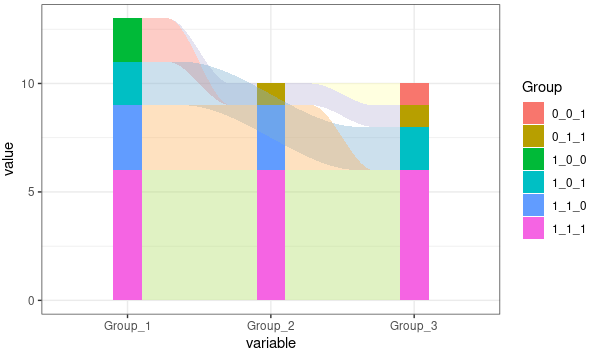
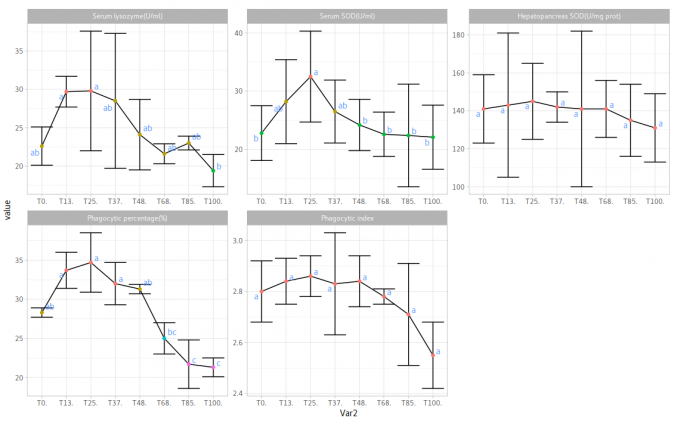
library(stringr)
library(ggplot2)
library(reshape2)
library(RColorBrewer)
Kaboom_Flow <- function(TB){
coul = brewer.pal(12, "Set3")
Group = c()
for(i in c(1:nrow(TB))){
Group= c(Group, paste(TB[i,],collapse = "_"))
}
Group_TB_gene = data.frame(Group,row.names = row.names(TB))
Group = as.data.frame(table(Group))
Group = Group[order(Group$Group, decreasing = F),]
Group_N = data.frame(str_split_fixed(Group[[1]], "_", ncol(TB)), stringsAsFactors =F)
for(i in c(1:ncol(Group_N))){
Group_N[[i]] = as.numeric(Group_N[[i]])
}
Group_N <- Group_N * Group$Freq
colnames(Group_N) = colnames(TB)
Increasing_list = Group_N
for(i in c((nrow(Increasing_list)-1):1)) {
Increasing_list[i,] = Increasing_list[i+1,] + Increasing_list[i,]
}
Increasing_TB <- rbind(Increasing_list, rep(0, ncol(Increasing_list),))
Group_N$Group <- Group$Group
Group_TB <- melt(Group_N)
Mutation_flow<- function(TB, Bar_w= 0.2){
P <- ggplot() +
geom_bar(data=Group_TB, aes(x=variable, y=value, fill = Group), stat = 'identity', position = 'stack', width = Bar_w) + theme_bw()
return(P)
}
Connect <- function(TMP_TB, Bar_w = 0.2, C_alp = .1, Color="grey"){
Indent_ = Bar_w/2
C_alp = (max(TMP_TB$X)- min(TMP_TB$X) -Bar_w) * C_alp + Indent_
TMP_TB2 <- TMP_TB
TMP_TB3 <- TMP_TB
TMP_TB2$X <- TMP_TB2$X + Indent_
TMP_TB3$X <- TMP_TB3$X - Indent_
TMP_ind <- rbind(TMP_TB2, TMP_TB3)
TMP_ind <- TMP_ind[which(TMP_ind$X %in% c(max(TMP_ind$X), min(TMP_ind$X))==FALSE),]
TMP_TB2$X <- TMP_TB2$X + C_alp
TMP_TB3$X <- TMP_TB3$X - C_alp
TMP_alp <- rbind(TMP_TB2, TMP_TB3)
TMP_alp <- TMP_alp[which(TMP_alp$X %in% c(max(TMP_alp$X), min(TMP_alp$X))==FALSE),]
Area_TB = rbind(TMP_ind, TMP_alp)
g1 <- ggplot() +
geom_smooth(data=Area_TB[Area_TB$line=="UP",], aes(x=X, y = value))+
geom_smooth(data=Area_TB[Area_TB$line=="DOWN",], aes(x=X, y = value))
gg1 <- ggplot_build(g1)
df2 <- data.frame(x = gg1$data[[1]]$x,
ymin = gg1$data[[1]]$y,
ymax = gg1$data[[2]]$y)
p <- geom_ribbon(data = df2, aes(x = x, ymin = ymin, ymax = ymax),
fill = Color, alpha = 0.4)
return(p)
}
Index_ = c()
for(Row in c(1:nrow(Group_N))){
TMP <- Group_N[Row,1:(ncol(Group_N)-1)]
Result = TMP[which(TMP!=0)]
tmp_id = which(colnames(TMP)==colnames(Result)[ncol(Result)])
if(tmp_id != ncol(TMP)){
Result[colnames(TMP)[tmp_id+1]] = 0
}
tmp_id = which(colnames(TMP)==colnames(Result)[1])
if(tmp_id != 1){
Result[colnames(TMP)[tmp_id-1]] = 0
}
Result = Result[as.character(sort(factor(colnames(Result),
levels = colnames(TMP))))]
row.names(Result) = Row
Index_ = c(Index_, list(Result))
}
P <- Mutation_flow(TB)
for(i in c(1:length(Index_))){
TMP = Index_[[i]]
Row = as.numeric(rownames(TMP))
TMP = Increasing_TB[Row:(Row+1),colnames(TMP)]
TMP$line=c("UP","DOWN")
for(col_i in c(1:(ncol(TMP)-2))){
TMP_TB = melt(TMP[c(col_i,col_i+1, ncol(TMP))])
TMP_TB$X = as.numeric(factor(TMP_TB$variable , levels=colnames(Increasing_TB
)))
P <- P + Connect(TMP_TB,.2, .1, coul[1+(i%%12)])
}
}
print(P)
return(list(P,Group_TB_gene))
}
|