Go Ontology Bar Plot by ggplot
Data
Before running the codes, I’d like to have a brief introduces about my data.
There are three tables in total and name as In_all.table, Li_all.table, Mu_all.table.
It looks like:
| Name | InCK_30 | InCK_75 | In30_75 | Cate |
|---|---|---|---|---|
| localization | 114 | 149 | 94 | Biological process |
| single-organism process | 285 | 357 | 225 | Biological process |
| metabolic process | 375 | 413 | 261 | Biological process |
| multi-organism process | 4 | 5 | 1 | Biological process |
| immune system process | 15 | 13 | 9 | Biological process |
| multicellular organismal process | 16 | 13 | 11 | Biological process |
| cellular process | 411 | 473 | 296 | Biological process |
| reproduction | 0 | 1 | 1 | Biological process |
| reproductive process | 0 | 1 | 1 | Biological process |
Quick Start
|
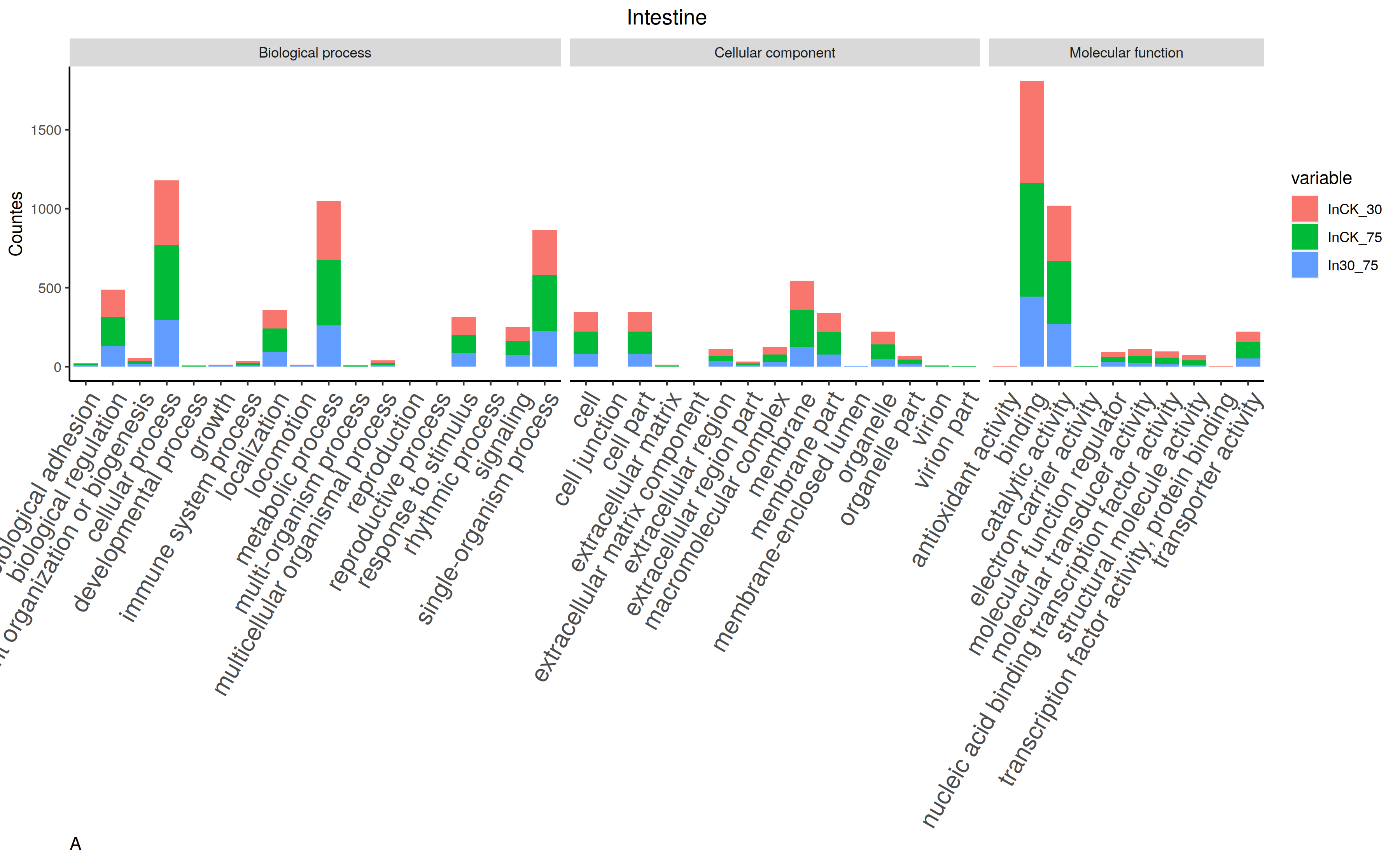
It is definitely the best result of GO bar plot.
But, thing gonna change when you want to align two or more others.
Plot Three files
|
Due to the name of categories are long and dense, it is hard to get an idea distribution of the chart.
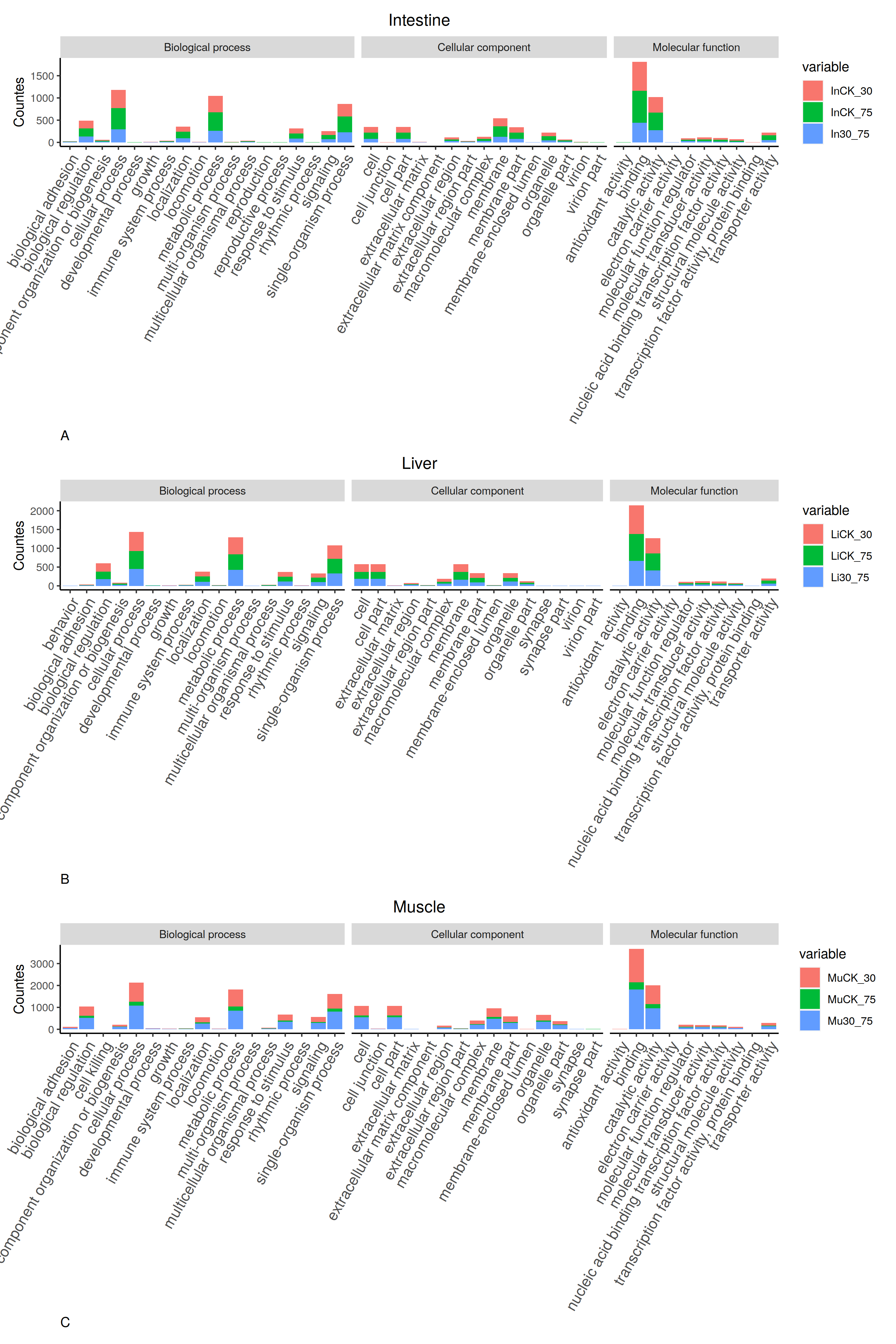
One resolution is to combine three tables and sharing one X axis. Another is to annotate the categories at the side of the chart.
Combining X axis
|
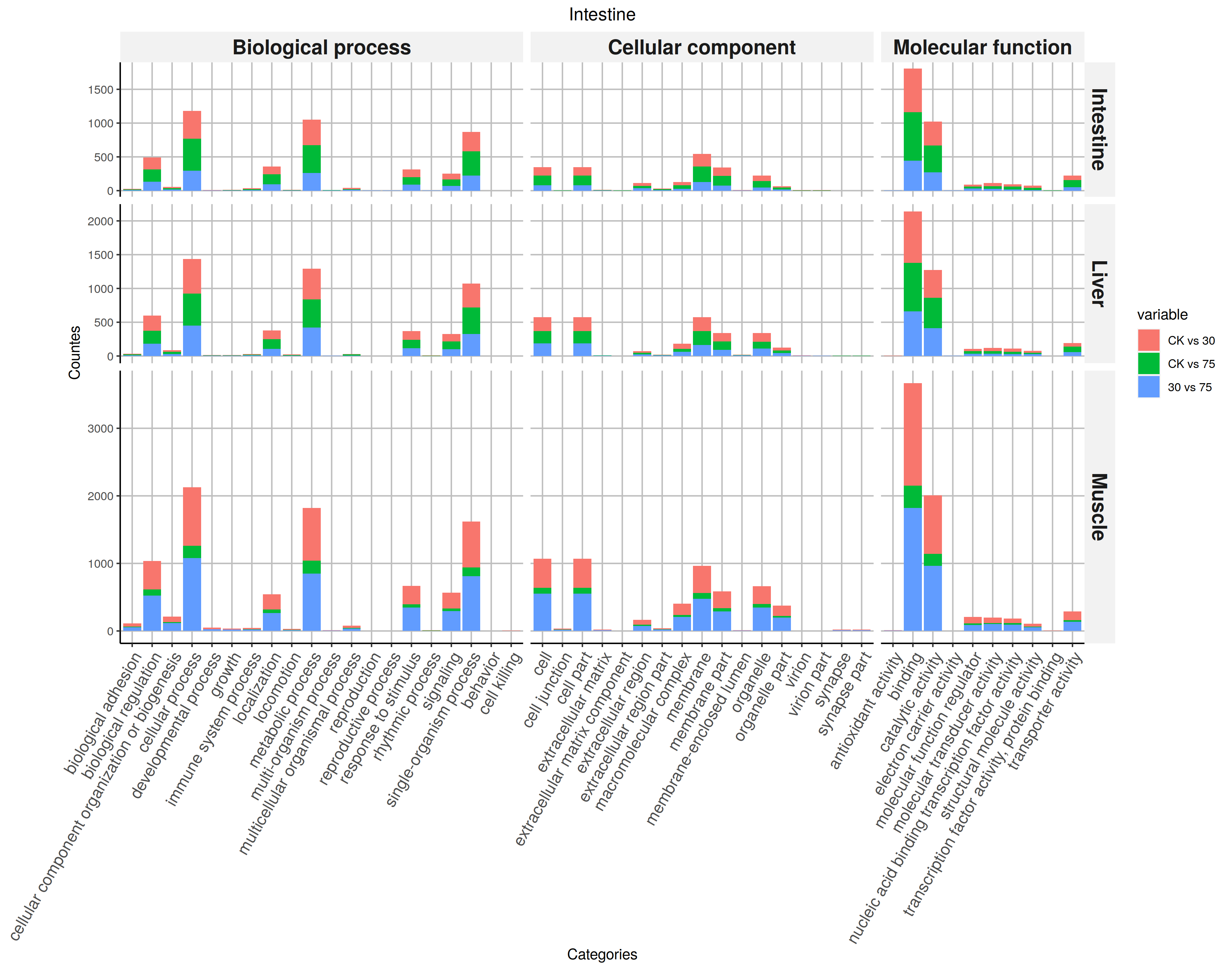
You may also like using heatmap to display the difference of counts among samples.
Heatmap
|
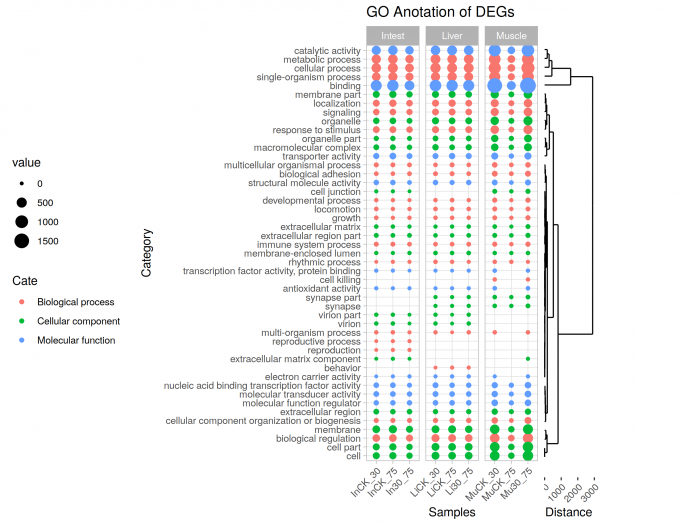
For have a better layout and easy to comparation, I tossed some legends.
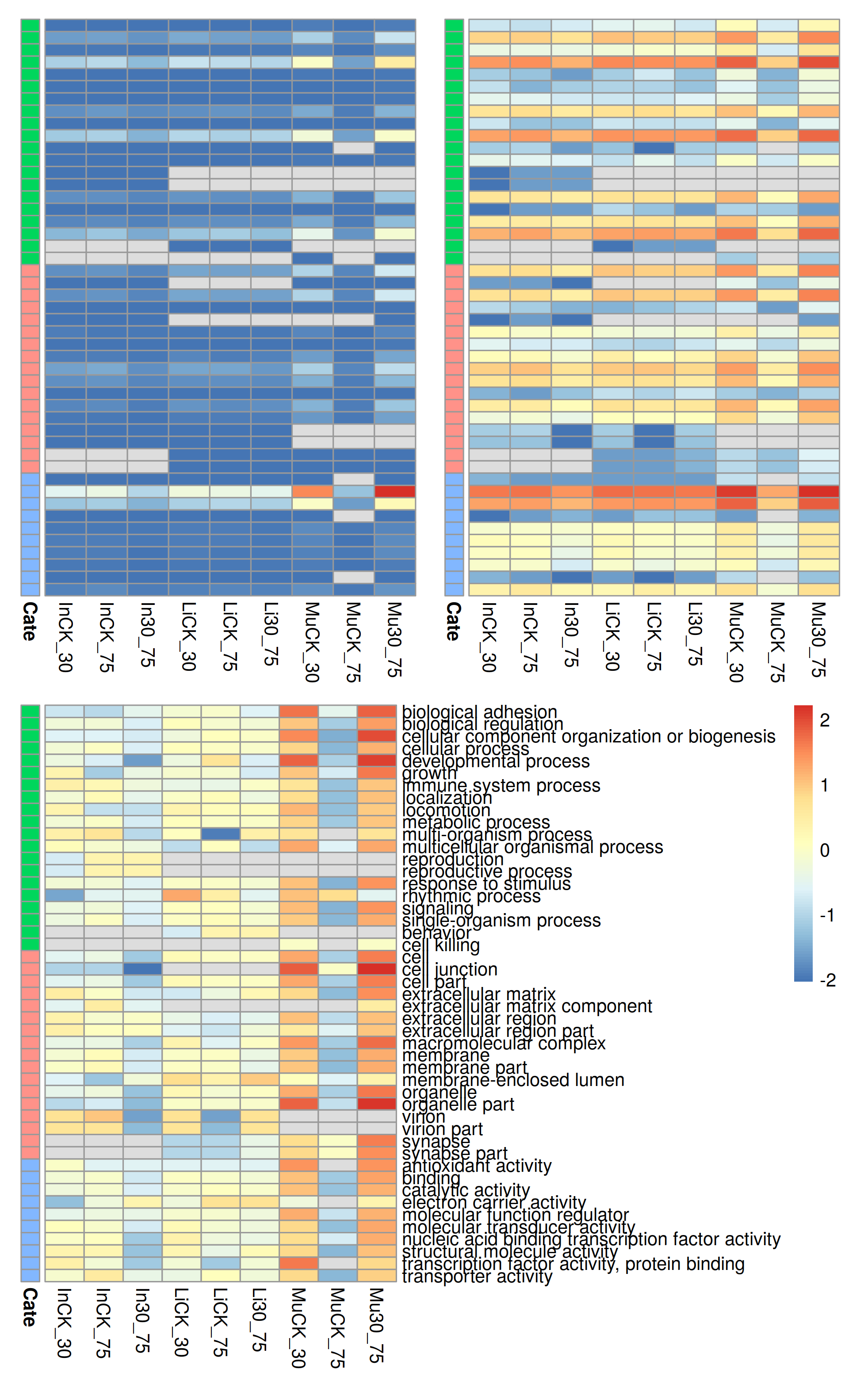
Dots plot
|
Go Ontology Bar Plot by ggplot







