刪除低map的reads
1. 讀取counts文件並篩選
1.1 頻率分佈統計
A <- read.csv("/media/ken/Data/Yan/RNA-seq/report/4.exprs//All_isoform.COUNT.matrix.anno.xls",sep='\t')
head(A)
|
X.ID1 ID2 Liver_CK Intest_CK Muscle_CK Liver_30 Intest_30 Muscle_30 Liver_75 Intest_75 Muscle_75
1 TRINITY_DN100000_c1_g1 TRINITY_DN100000_c1_g1_i1 212 331 89 128 401 223 310 407 130
2 TRINITY_DN100001_c0_g1 TRINITY_DN100001_c0_g1_i1 35 1244 1 14 609 261 12 1085 0
3 TRINITY_DN100002_c0_g1 TRINITY_DN100002_c0_g1_i1 27 4 2 38 7 3 19 16 7
4 TRINITY_DN100002_c1_g1 TRINITY_DN100002_c1_g1_i1 23 2 34 4 6 18 8 5 51
5 TRINITY_DN100002_c2_g2 TRINITY_DN100002_c2_g2_i1 0 1 0 0 0 0 1 1 0
6 TRINITY_DN100002_c2_g3 TRINITY_DN100002_c2_g3_i1 0 0 40 0 0 62 0 1 84
|
row.names(A)=A[[2]]
A$Sum = rowSums(A[-c(1:2)])
|
1.2 繪圖
library(ggplot2)
library(reshape2)
ggplot(A[c("ID2","Sum")])+ geom_density(aes(x=Sum)) + xlim(c(0,25)) + theme_linedraw()
|
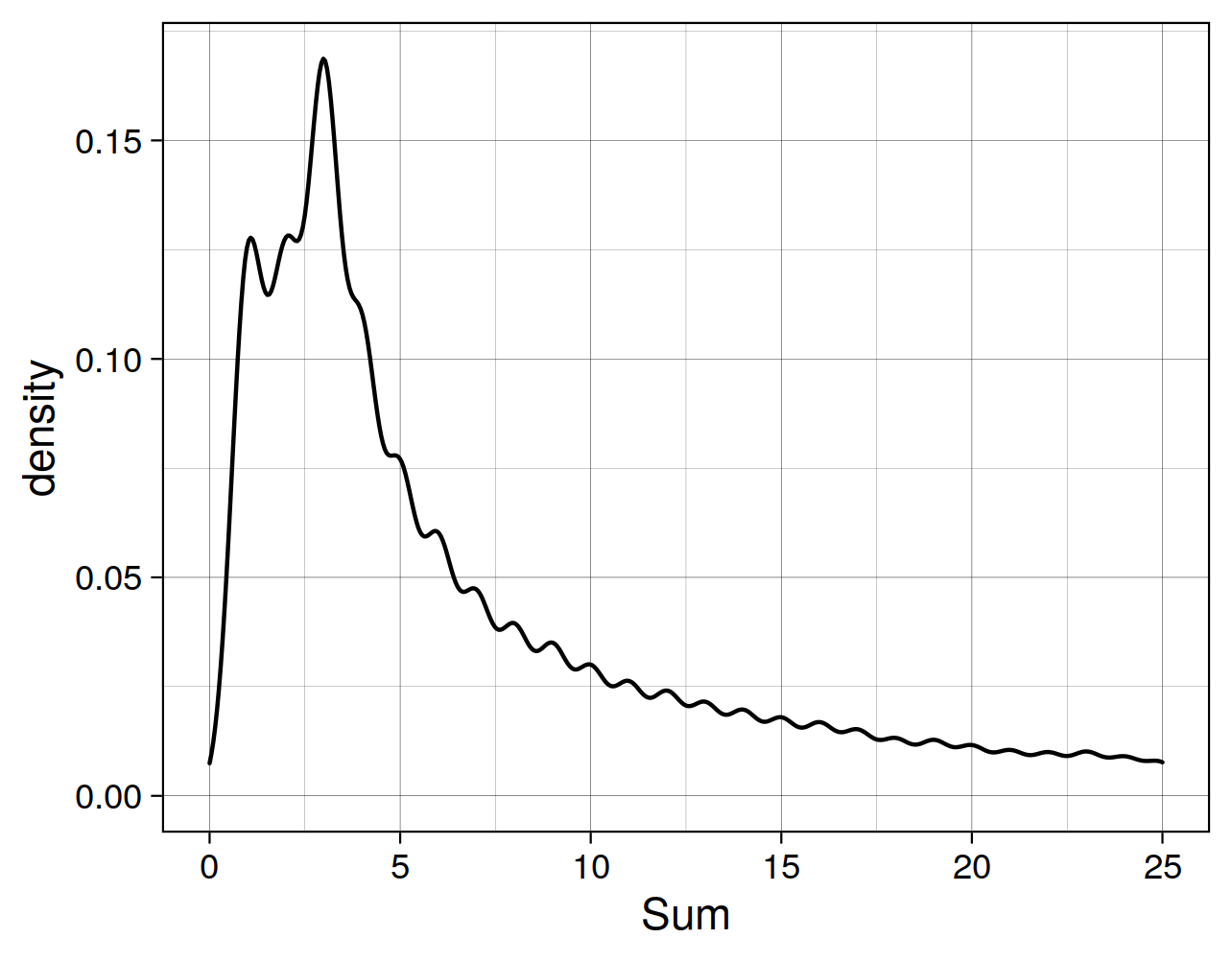
如圖所示, 大部分的hit總和小於等於3。應爲有9個樣本, 所以我們把閾值設置在10,則:
A_sub = A[which(A$Sum>10),]
paste(round((nrow(A_sub)/nrow(A))*100,2),"%",sep="")
|
可知, 剩餘"52.86%"的reads被保存了
輸出ID:
write.table(A_sub[[2]],"filt.list",row.names = F,quote=F,col.names=F)
|


