KEGG result visualization by ggplot
KEGG result visualization by ggplot
用ggplot画KEGG结果
Data
Before running the codes, I’d like to have a brief introduces about my data.
As you can see codes below, List has all files I need.
Take Intest_30_vs_Intest_75 as an example:
| #KEGGid | KEGGdescription | KEGGclass | KEGGsubclass | Oddsratio | p-value | q-value | Gene_numbers |
|---|---|---|---|---|---|---|---|
| 04610 | Complement and coagulation cascades | Organismal Systems | Immune system | 11.7158541941 | 6.30141083687e-10 | 1.18731846295e-07 | 14 |
| 04977 | Vitamin digestion and absorption | Organismal Systems | Digestive system | 11.1739766082 | 0.000269773611998 | 0.0254155139724 | 5 |
| 04974 | Protein digestion and absorption | Organismal Systems | Digestive system | 3.73319892473 | 0.000445170135803 | 0.0279598085294 | 11 |
Quick Start
|
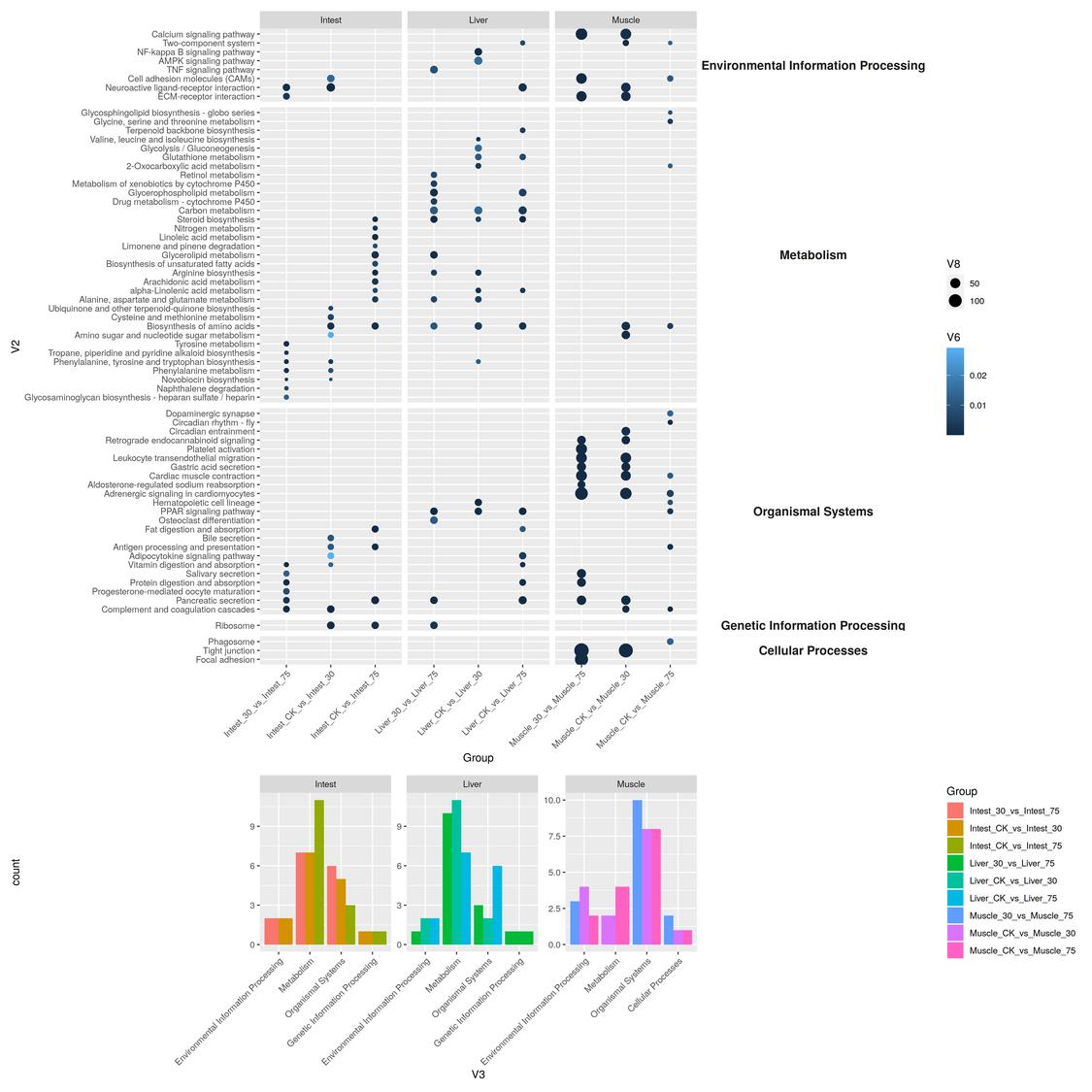
More complicate plot
Functions TreePlot and LS_judg could be found at: Blog/Yueque
|
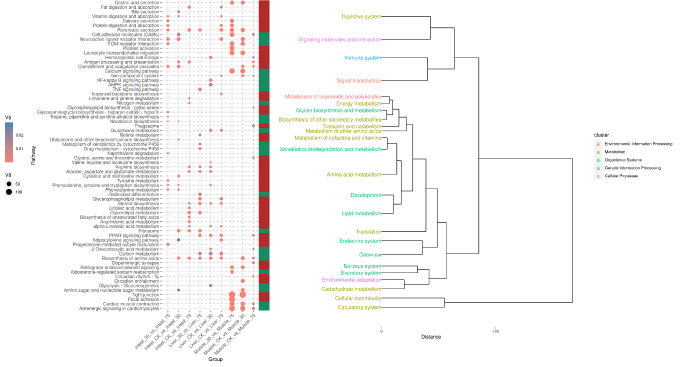
More
Get a KEGG results first
codes from: ClusterProfiler
|
$Cos\alpha = \frac{tA}{tC}$
$Sin\alpha = \frac{\sqrt{tC^2 - tA^2}}{tC}$
$mC = \frac{mB_1}{Sin\alpha}$
$mtC = mC * \frac{mB_1 - mB_2}{mB_2}$
$S = mtC/tC$
$S = \frac{mC * \frac{mB_1 - mB_2}{mB_2}}{tC}$
$S = \frac{\frac{mB_1}{Sin\alpha} * \frac{mB_1 - mB_2}{mB_2}}{tC}$
$S = \frac{\frac{mB_1}{\frac{\sqrt{tC^2 - tA^2}}{tC}} * \frac{mB_1 - mB_2}{mB_2}}{tC}$
$S = \frac{\frac{mB_1}{\frac{\sqrt{tC^2 - tA^2}}{tC}} * \frac{mB_1 - mB_2}{mB_2}}{tC}$
$S = \frac{\frac{mB_1^2 - mB_1 * mB_2 }{\frac{mB_2\sqrt{tC^2 - tA^2}}{tC}}}{tC}$
$S = \frac{mB_1^2 - mB_1 * mB_2 }{\frac{mB_2\sqrt{tC^2 - tA2}}{tC2}}$
$S = \frac{mB_1^2 - mB_1 * mB_2 }{tC^2 * mB_2\sqrt{tC^2 - tA^2}}$
KEGG result visualization by ggplot