Linear Regression
Regression in R
Reference: Multiple linear regression made simple; 2021
linear equation with one unknown
test data: mpg form ggplot2
Quick start
|
Call:
lm(formula = hwy ~ displ, data = mpg)
Coefficients:
(Intercept) displ
35.698 -3.531
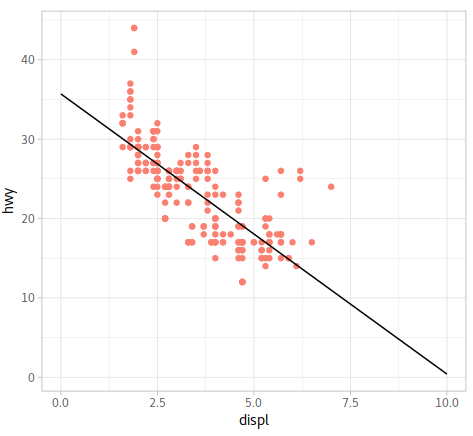
As we can see above, the function is
$$
y = -3.531x+35.698
$$
|
Call:
lm(formula = hwy ~ displ, data = mpg)
Residuals:
Min 1Q Median 3Q Max
-7.1039 -2.1646 -0.2242 2.0589 15.0105
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 35.6977 0.7204 49.55 <2e-16 ***
displ -3.5306 0.1945 -18.15 <2e-16 ***
---
Signif. codes: 0 ‘**\*’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 3.836 on 232 degrees of freedom
Multiple R-squared: 0.5868, Adjusted R-squared: 0.585
F-statistic: 329.5 on 1 and 232 DF, p-value: < 2.2e-16
Plot
|
Practice
P value and R2
|
Call:
lm(formula = n ~ m, data = df)
Coefficients:
(Intercept) m
-0.01162 0.01353
Call:
lm(formula = n ~ m, data = df)
Residuals:
Min 1Q Median 3Q Max
-0.020694 -0.006615 -0.001036 0.005432 0.026901
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -0.011621 0.008968 -1.296 0.224
m 0.013531 0.001219 11.105 6.04e-07 ***
---
Signif. codes: 0 ‘\***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 0.01457 on 10 degrees of freedom
Multiple R-squared: 0.925, Adjusted R-squared: 0.9175
F-statistic: 123.3 on 1 and 10 DF, p-value: 6.036e-07
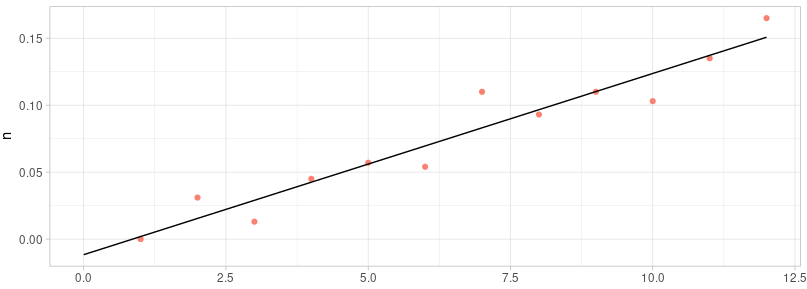
We can know that the function of this result is:$y=0.01353x-0.01162$
The R2 = 0.925, p-value = 6.036e-07
According to the P-value, we could say that the x and y are significantly correlated. Based on the R2, we can say the fitting result is pretty reliable.
To be notice that for the slope, the sd is 0.00012 and for the intercept, the sd is 0.0090. They are independent.
Turn R to P
There are two online tools which can calculate the P value from R (not R2):
And there are also code for calculate the p value from R2:
Reference: Jake Westfall; 2014
The formula is:
$$
r^ 2 ≈ Beta(\frac{V_ n}{2}, \frac{V_ d}{2})
$$
Vn and Vd are the numerator and denominator degrees of freedom, respectively, for the associated F-statistic.
|
Runs test
|
Runs Test for Randomness data: TMP runs = 8, m = 7, n = 5, p-value = 0.8535 alternative hypothesis: true number of runs is less than expected
This runs test for randomness is achieved by divided the raw data into two groups, the elements larger than the predicted values are assigned into 1, smaller elements are assigned into -1. After that, we only use the “less” part the calculate the p value and find which is not significant. So, we can say that they are not significant different and the regression function is acceptable.
PS: this is are exactly Prism suppose to run the runs test
Confidence and prediction intervals are often formed to answer questions such as the above. Intervals allow one to estimate a range of values that can be said with reasonable confidence (typically 95%) contains the true population parameter. It should be noted that although the questions above sound similar, there is a difference in estimating a mean response and predicting a new value. This difference will be seen in the interval equations. This post will explore confidence and prediction intervals as well as the confidence ‘bands’ around a linear regression line.
rpubs.com: Confidence Interval of the linear regression
|
Outliers
 |
|---|
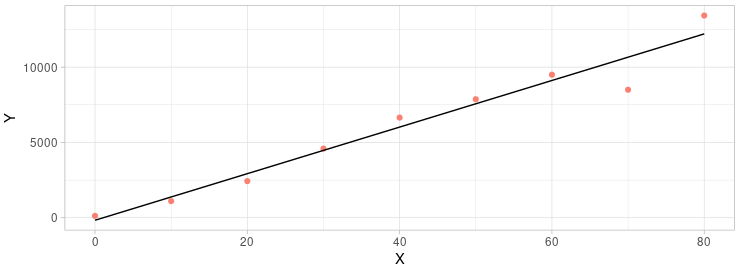
To check the outliers we could use the chauvenet’s test which is based on the t-test. The basic idea is to check the t-distribution of the difference between the real number and the expected number. The number is different from the expected and can’t pass the chauvenet’s test been seam as outliers.
As you can see from the graph above (the code for the plot is attached below), the last two points are far from the regression line. Mandatory discarding both of them would be arbitrary and inconvincible. But it would be a good idea to use the chauvenet’s test to decide which one should be kept and which one should be discarded.
|
Call:
lm(formula = Y ~ X, data = TB)
Residuals:
Min 1Q Median 3Q Max
-2164.1 -278.1 289.6 383.6 1218.2
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -169.56 624.28 -0.272 0.794
X 154.77 13.11 11.803 7.1e-06 ***
---
Signif. codes: 0 ‘\***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 1016 on 7 degrees of freedom
Multiple R-squared: 0.9522, Adjusted R-squared: 0.9453
F-statistic: 139.3 on 1 and 7 DF, p-value: 7.105e-06
Though the P-value and R2 tell us that the fitness result is very promising, we still want to see how the outlier affects the fitness.
|
The output of the chauvenet_test
[1] "Dmax: 1.91450582505556" [1] "Outliners: -2164.34" [1] "Zscore: 2.27790206887637" [1] "P value: 0.380218526950083" "P value: 0.384907967278372" [3] "P value: 0.300921524723487" "P value: 0.451166958976122" [5] "P value: 0.254009368820307" "P value: 0.37561761649201" [7] "P value: 0.343240779142229" "P value: 0.0113662064986361" [9] "P value: 0.0999076632824158" [1] -2164.34
|
[1] 8
We can see the the 8th element in Y could be discarded. Let’s we run everything again without it.
|
Call:
lm(formula = Y ~ X, data = TB[-8, ])
Residuals:
Min 1Q Median 3Q Max
-528.3 -186.7 -113.3 304.4 549.9
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -429.900 239.097 -1.798 0.122
X 169.411 5.432 31.188 7.22e-08 ***
---
Signif. codes: 0 ‘\***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 383.6 on 6 degrees of freedom
Multiple R-squared: 0.9939, Adjusted R-squared: 0.9928
F-statistic: 972.7 on 1 and 6 DF, p-value: 7.217e-08
 |
|---|
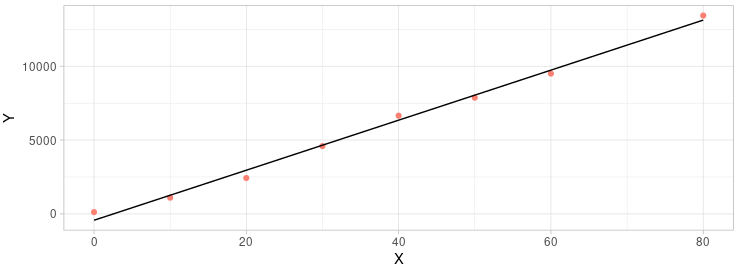
We can see that the fitness was much better than before when we discard the 8th element. The R2 is larger than before and P-value is smaller than before. Seams we had a better result than before.
Confidential intervals
There is two way you can do for draw confidential intervals. One is using ggplot2. The functions geom_smooth could help you finish it very elegantly. Another way is to follow the steps from rpubs which calculate it by yourself.
In ggplot
Let’s see how can we achieve it with ggplot2. The threshold of the interval is 0.95 which could be changed in level =
|
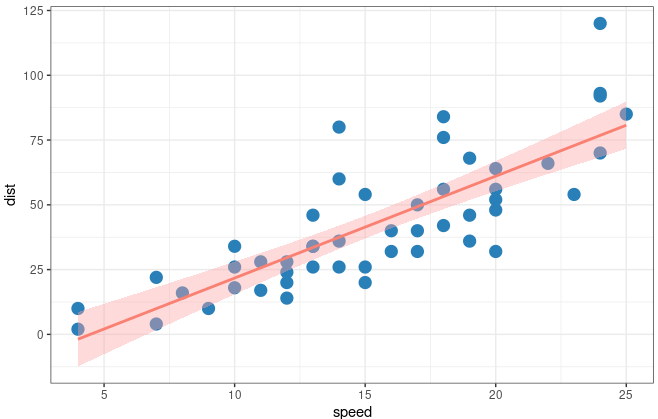 |
|---|
Calculating confidence interval by yourself:
Points from upper and lower side would be calculated and stored.
|
Lower Confidence Band Upper Confidence Band 1 -33.350170 6.056798 2 -29.417761 9.989207 3 -21.327925 9.764188 4 -17.395516 13.696597 5 -12.154300 16.320197 6 -6.971285 19.002000
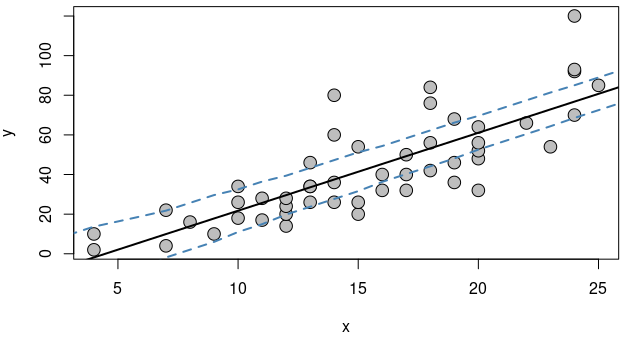 |
|---|
Quadratic Equation
Standard Equation:
$$
y=ax^2+bx+c
$$
|
model_mpg1:
Call:
lm(formula = hwy ~ I(displ^2), data = mpg)
Coefficients:
(Intercept) I(displ^2)
29.324 -0.429
model_mpg2:
Call:
lm(formula = hwy ~ I(displ^2) + displ, data = mpg)
Coefficients:
(Intercept) I(displ^2) displ
49.245 1.095 -11.760
plot
|
| model_mpg1 | model_mpg2 | |
|---|---|---|
| Formula | $y=-0.429x^{2}+29.324$ | $y=1.095x^{2} -11.760x + 49.245$ |
| Plot | 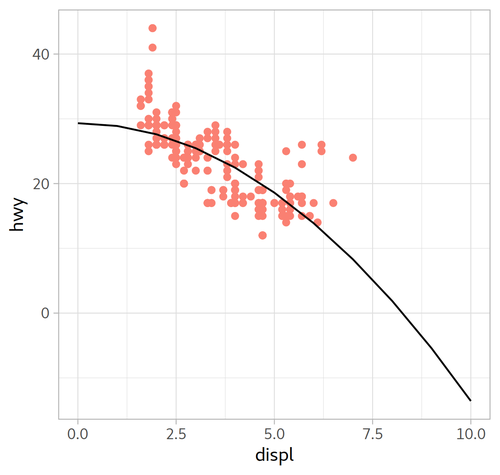 |
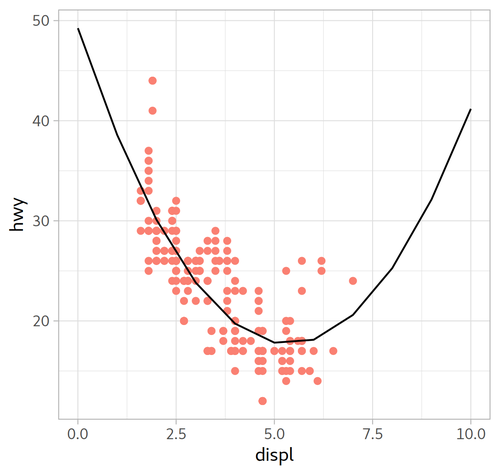 |
|
R2
|
一元多次回归方程自动扫描
T test and P value: 数据挖掘运爷
Functions:
Click to see
|
Usage:
|
| Exp | P_value | R2 | F | |
|---|---|---|---|---|
| ssr | 1 | 5.100486e-02 | 0.9641461 | $40.2380952380953 + 4.78571428571428 * X$ |
| 1 | 2 | 5.100486e-02 | 0.9865977 | 43.5714285714286+0.250000000000001 * X^2+2.28571428571428 * X^1 |
| 11 | 3 | 1.550240e-08 | 0.9986254 | 41.904761904762+0.069444444444444 * X^3±0.791666666666658 * X^2+6.0912698412698 * X^1 |
| 12 | 4 | 3.012296e-02 | 0.9986254 | 41.904761904762+2.01944464079605e-16 * X^4+0.0694444444444399 * X^3±0.791666666666634 * X^2+6.09126984126975 * X^1 |
| 13 | 5 | NaN | 1.0000000 | 41.9999999999999±0.00625000000000004 * X^5+0.156250000000001 * X^4±1.29166666666667 * X^3+3.99999999999998 * X^2+0.516666666666805 * X^1 |
Logarithmic Regression
|
Call:
lm(formula = hwy ~ log(displ, exp(1)), data = mpg)
Coefficients:
(Intercept) log(displ, exp(1))
38.21 -12.57
|
drc
|
Test:
Linear Regression








