Principles of Biochemistry 8 |Enzyme Design| Class Notes |HarvardX
The enzymes were used in different areas to fit different purposes. But there are so many restrictions before applying them in utility. As a result, enzyme modification is a huge leap for the enzyme industry.
Tree enzyme design approaches:
Rational Design
Using existing structural information of the enzyme, and using a rational approach to replace specific amino acids with others.
It is based on the structural information and another ligand for the enzyme.
Drawbacks:
- Full structural information of the enzyme with and without substrate is required.
- Time-consuming
- Money
- Complexity
Directed by Evolution
Evolution on a very small scale in a controlled environment.
White type protein;
introducing a variety of mutations;
Screen and select the result;
back to the first step and mutated it again.
Drawbacks:
- Not all enzymes are amenable to high-throughput screens.
Semi-rational Design
Iterative Saturation Mutagenesis (ISM)
- the selected sites are saturated with mutations.
- saturation mutagenesis is repeated to obtain an optimum variant
Saturation Mutagenesis
- Mutation improves activity or modifies the specificity of enzymes located at, or near, the active site.
- The mutation improves the stability of the enzyme tend to away from the activity site.
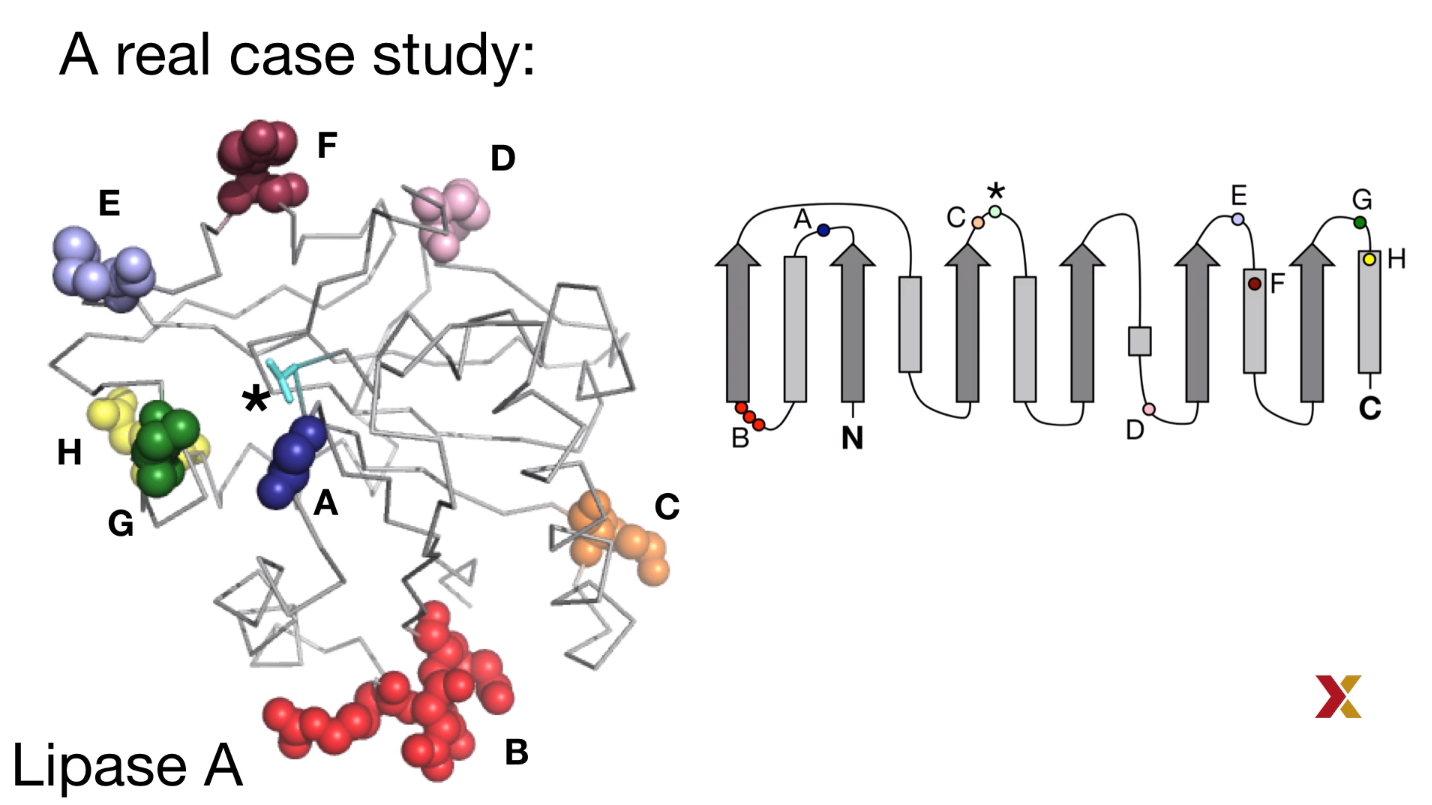
Example: lipase A
T50: the temperature at which 50% of the enzyme remains properly folded and active after a given time interval.
-
B-factor which could measure the flexibility of amino acid was applied, and the top 10 highest B-factor AA at 8 sites in wild-type structure were selected.
-
Hypothesis: the mutation of these sites could increase the rigidity of the protein and wouldn’t affect the activity of the enzyme.
| © HarvardX |
|---|
 |
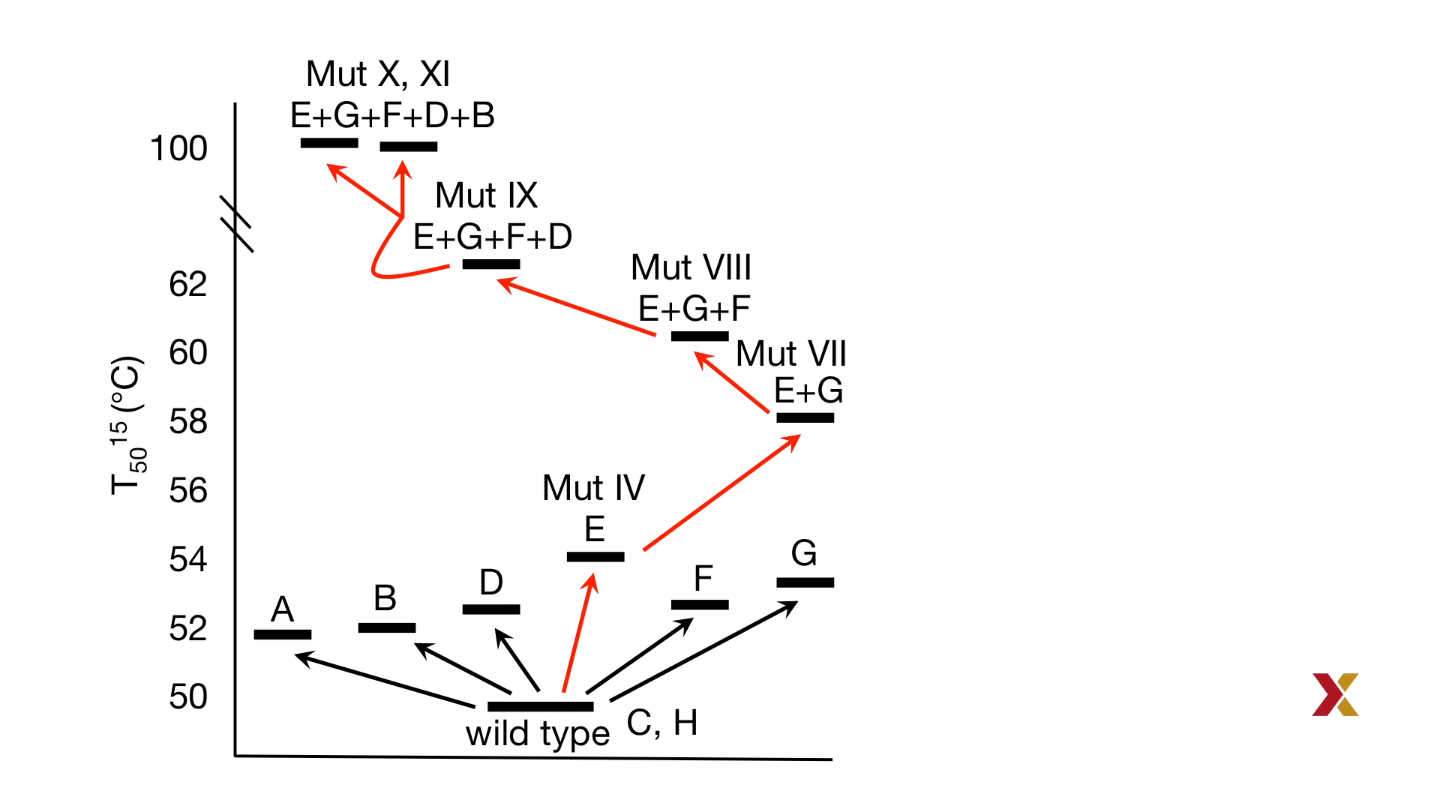 |
Result:
Condition: pre-incubation at $75^ \circ C$ for $60 min$
| Protein | Residual Activity |
|---|---|
| WT | 0% |
| Variant X | 60% |
| Variant XI | 90% |
ISM applications
Iterative Saturation Mutagenesis:
- Crystal structure -> choose AA
- ‘Smart’ mutant library
- Assay enzyme activity
- Repeat
Universal Blood
Back ground:
Universal blood is the blood, which could be used for any of the patients.
AB blood type system: the different sugar groups found on glycolipids on the surface of the red blood cell.
They have a glycoside hydrolase that cleaves the antigenic trisaccharides found in type A and B red blood cells.
Goals: improve the activity of glycoside hydrolase
- Figure out the crystal structure of the enzyme
- Generate mutant library by degenerate-code PCR
- Screen for enzyme activity to cleave fluorescent glyco-substrate.
Result: By repeating screen and selection, scientists finally replaced 6 AAs and increased 170-fold of activity compared to the wild type.
Halohydrin dehydrogenase
manufacture of pharmaceutical compounds.
Background:
Challenge: The production of the drug is often a mixture of stereoisomers.
epoxide group: a common precursor in chemical synthesis which is very active.
Normal circumstance:
Halohydrin dehydrogenase + R-2-chloro-1-phenylethanol -> pure product.
S-2-chloro-1-phenylethanol: remains no change.
Solution: Design an enzyme that could tolerate the S form of the substrates.
- Solving the crystal structure and determine the (7) amino acids in activity sites.
- Generate mutant enzyme library by degenerate codon PCR
- High throughput colorimetric screen for enzyme activity.
Result: As a result, the combination use of the wild and mutation type enzyme was used to convert both forms of the substrates to the desired product.
Diabetes sensors
Background:
Glucometers: convert glucose into a product and release electrons.
Naturally, oxygen would accept electrons and lead to underestimating the level of glucose.
Goals: Increasing the accuracy of the Glucometers.
-
Target glucose oxidase amino acid sites were chosen based on functional information.
-
build a mutagenesis lib:
- alanine 173 & 332 increases both mediator and oxygen activities.
- phenylalanine 414, increased mediator activity, and decreased oxygen activity.
- valine 560 dramatically decreased oxygen activity.
-
Muti-site saturation mutagenesis to find optimal oxygen-insensitive enzymes.
Result: Variant 7
| Wilde | Position | Mutate |
|---|---|---|
| Adenine | 173 | valine |
| Alanine | 332 | serine |
| Phenylalanine | 414 | isoleucine |
| Valine | 560 | threonine |
Alter the use of the cofactor
Target: Increased the by-path method of reaction.
- The crystal structure was resolved and the residuals near the active site were determined.
- Site-saturation mutagenesis via PCR with NNK degenerate primers at the 54 positions.
- High throughput screens identified mutations at 12 sites that boosted NAD(H) usage.
- The best variant, Q350N was used as the template for ISM for the remaining 11 positions.
- An additional variant, Q350N/H171A, was discovered with further increased NAD(H) usage.
Principles of Biochemistry 8 |Enzyme Design| Class Notes |HarvardX
https://karobben.github.io/2021/04/11/LearnNotes/edx-biochm-8/








