Plot a network
R library(igraph)1 )0 :1 , 100 , replace=TRUE , prob=c (0.8 ,0.2 )), nc=10 )'undirected' , diag=F )c (0 ,0 ,0 ,0 ))
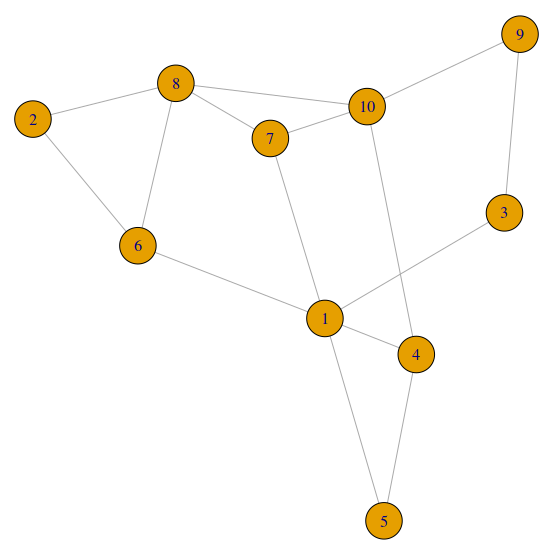
This is the data structure we’d like to have
[,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8] [,9] [,10]
[1,] 0 0 1 0 1 0 1 0 0 0
[2,] 0 0 0 0 0 1 0 1 0 0
[3,] 0 0 0 0 0 0 0 0 0 0
[4,] 1 0 0 0 0 0 0 0 0 1
[5,] 0 0 0 1 0 0 0 0 0 0
[6,] 1 0 0 0 0 0 0 1 0 0
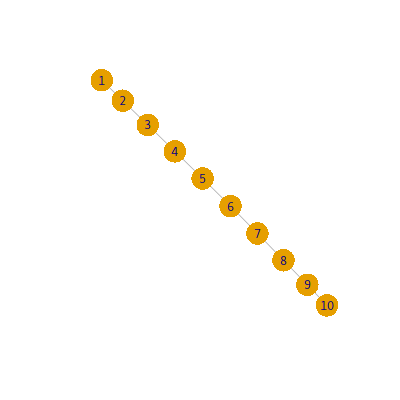
From List to Network-Data Matrix
Let’s say, a list of data 1, 2, 3, 4 means the relationship of 1-> 2 -> 3 -> 4.
R List <- c (1 , 2 , 3 , 4 , 5 , 6 , 7 , 8 , 9 , 10 )function (List){c (List[-1 ],NA )"A" =List, "B" = tmp, V = 0 )c ("A" , "B" , "V" )return (na.omit(TB))function (TB){'A' ,idvar='B' , direction='wide' )c (matrix(t(data.frame(strsplit(colnames(AA),"[.]" )))[,2 ]))1 ]1 ]1 c (colnames(AA), rownames(AA)))is.na (AA)]=0 return (AA)
1 2 3 4 5 6 7 8 9 10
1 0 1 0 0 0 0 0 0 0 0
2 0 0 1 0 0 0 0 0 0 0
3 0 0 0 1 0 0 0 0 0 0
4 0 0 0 0 1 0 0 0 0 0
5 0 0 0 0 0 1 0 0 0 0
6 0 0 0 0 0 0 1 0 0 0
7 0 0 0 0 0 0 0 1 0 0
8 0 0 0 0 0 0 0 0 1 0
9 0 0 0 0 0 0 0 0 0 1
10 0 0 0 0 0 0 0 0 0 0
R network=graph_from_adjacency_matrix(data.matrix(AA), mode='undirected' )"network.png" ,sep="" ), width = 400 , height = 400 )1 ,0.7 ,"" ,"transparent"
© Karobben
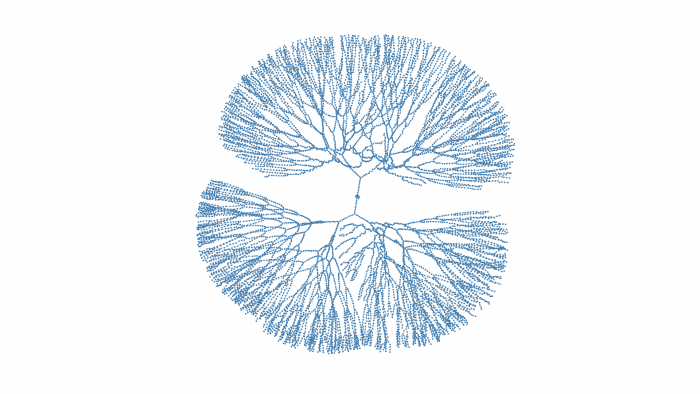
For loop to build a list
Tree_grwoth <- function (Growth_Turn = 10 , Period = 2 ){for (f in c (2 :Growth_Turn)){c ("A" )list ("main" = Main)for (t in c (1 :Turns)){for (branch in names (Result)){c ( Result[[branch]], paste( Result[[branch]][1 ],t, sep="-" ))if (length (branch_tmp) %% Period == 0 ){"New Branch" )length (branch_tmp)]] = c (branch_tmp[length (branch_tmp)])return (Result)20 , 5 )
Length Class Mode
main 21 -none- character
A-4 17 -none- character
A-4-8 13 -none- character
A-9 12 -none- character
A-4-8-12 9 -none- character
A-4-13 8 -none- character
$main
$main
[1] "A" "A-1" "A-2" "A-3" "A-4" "A-5" "A-6" "A-7" "A-8" "A-9" "A-10" "A-11" "A-12" "A-13" "A-14" "A-15"
[17] "A-16" "A-17" "A-18" "A-19" "A-20"
$`A-4`
[1] "A-4" "A-4-5" "A-4-6" "A-4-7" "A-4-8" "A-4-9" "A-4-10" "A-4-11" "A-4-12" "A-4-13" "A-4-14" "A-4-15"
[13] "A-4-16" "A-4-17" "A-4-18" "A-4-19" "A-4-20"
$`A-4-8`
[1] "A-4-8" "A-4-8-9" "A-4-8-10" "A-4-8-11" "A-4-8-12" "A-4-8-13" "A-4-8-14" "A-4-8-15" "A-4-8-16" "A-4-8-17"
[11] "A-4-8-18" "A-4-8-19" "A-4-8-20"
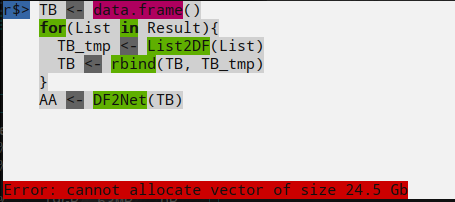
Whole Pipeline
Result <- Tree_grwoth(100 , 10 )'' ' STATE <- data.frame(summary(Result)) STAT <- STATE[STATE$Var2 == "Length",] sum(as.numeric(str_remove_all(as.character(STAT$Freq)," ")) -1) ' '' for (List in Result){'undirected' )1 ,0.7 ,"" ,"transparent"
When I try Result <- Tree_grwoth(135, 10):
V(network)$color ="steelblue" for (i in c (1 :100 )){".png" ,sep="" ), width = 1920 , height = 1080 )1 / tanh (0.004 * i),0.7 ,"" ,"transparent"