Amino Acid|Graduate Biochemistry 2| Tulane
Overview
Protein Structure
- Proteins are polymers of amino acids
- Native proteins are folded into a unique three dimensional structure
- The three dimensional structure is responsible for specificity and biological activity
- The structure is determined entirely by the primary sequence of amino acids through the physical & chemical properties of the amino acids
Amino Acid Properties
- Chemical Structure
- R group is variable
- 20 amino acids are found in proteins
- Amino acids are chiral (optically active)
- Amino acids in proteins always have the L configuration (Equivalent to the configuration of glyceraldehyde that rotates polarized light to the left)
- Levorotatory – Leftward rotation of light
- Dextrorotatory – Rightward rotation
- L amino acids are not all Levorotatory
20 AA
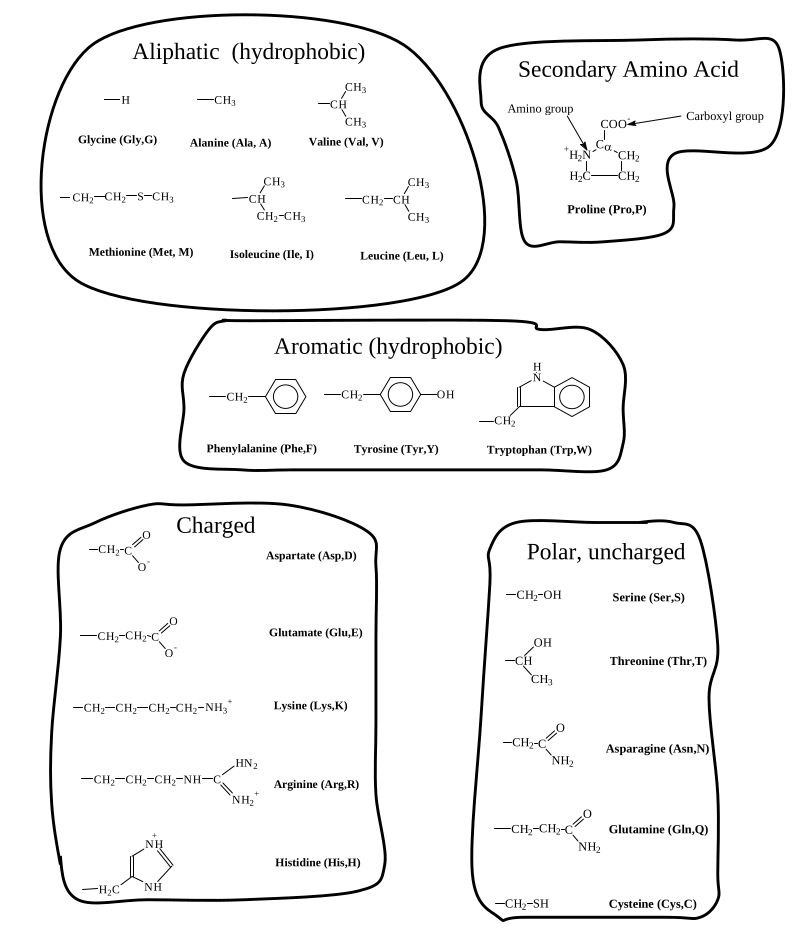
Remember the abb. and name and groups of all amino acids
| Gly | G | Glycine | Glycine is in the β turn |
| Ala | A | Alanine | Every where -> hydrophobic but not really strong |
| Val | V | Valine | hydrophobic, strictly in shape |
| Leu | L | Leucine | … |
| Ile | I | Isolucine | β branched |
| Met | M | Methionine | containing sulfate, hydrophobic |
| Pro | P | Proline | tight turn |
| Phe | F | Phenylalanine | |
| Tyr | Y | Tyrosine | Phenylalanine-(OH) |
| Trp | W | Tryptophan | largest hydrophobic group |
| Asp | D | Aspartate | β Carboxyl |
| Glu | E | Glutamate | γ Carbocyl |
| Lys | K | Lysine | ε Amino Group |
| Arg | R | Arginine | γ Guanidino Group |
| His | H | Histidine | β Imidazole Group |
| Ser | S | Serine | &beta hydroxyl |
| Thr | T | Threonine | hydroxyl gorup |
| Asn | N | Asparagine | Amide Group; Can not accept proton |
| Glu | Q | Glutamine | Amide Group; Can not accept proton |
| Cys | C | Cysteine |
Properties:
- Aliphatic (hydrophobic)
- Secondary Structured
- Aromatic (hydrophobic)
- Charged
- Polar, uncharged
Ionization Properties of Amino Acids
| AA | Function Group | pKa |
|---|---|---|
| Asp | -CH2-COO- | 3.9 |
| Glu | -CH2-CH2-COO- | 4.3 |
| Lys | -CH2-CH2-CH2-CH2-NH3+ | 10.8 |
| Arg | -CH2-CH2-CH2NH-C(-NH2)=NH2+ | 12.5 |
| His | -CH2-Imidazole | 6.5 |
primary structure
- Direction: N -> C
- Average molecule weight of per amino acid is ~110.
Properties of the Peptide Bond
- Electronic resonance gives the central -C(=O)N(H)- atoms some double-bond character
- Double bond character gives these bonds a generally planar (not tetrahedral) shape and rigidity
- Coplanarity severely limits the number of accessible conformations
Cis-trans isomerization
- Trans peptide bonds are energetically preferred
- Cis peptide bonds are rare:
- 0.05% of all non-proline peptide bonds are cis
- 6.5 % of all X-Pro peptide bonds are cis
- Rate of conversion between cis and trans is slow
Trans: opposite
Cis: same side
Conformational Properties of Polypeptides
- Protein backbone conformation can be described with 2 torsion angles Phi (Φ) and Psi (Ψ) around Cα
- Steric clashes make only some combinations of Φ and Ψ permissible
- The requirement for hydrogen bonding between backbone groups in folded proteins further limits the observed values for Φ and Ψ
| $\phi$ | $\psi$ |
|---|---|
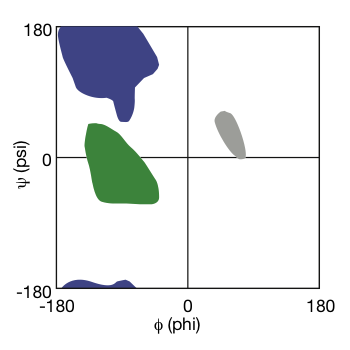 |
|---|
| Ramachandran Plot © HarvardX |
 |
|---|
| © Sepp Hochreiter |
Secondary Structure of Polypeptides
- Hydrogen Bonds are weak noncovalent interactions between polar groups.
- In a folded protein, backbone groups are always involved in hydrogen bonds.
- Hydrogen bonding along with the allowed φ , ψ torsion angles determine the possible types of secondary structure
- Secondary structure is the local conformation the backbone
- Secondary structure is defined by Hydrogen bonding patterns and Φ , Ψ torsion angles
α helix
 |
|---|
| © PDB ID=1SI4 |
- Right handed helix
- Interactions are local
- Defined by Hydrogen-bonding pattern (ith) C=0 - - - NH (i+4)th
- Accounts for well more than half of all protein structure
- Pitch: 3.6 residues per turn
- Rise: 1.5 Å per residue (5.4Å/turn)
- 13 atoms in the hydrogen bonded loop
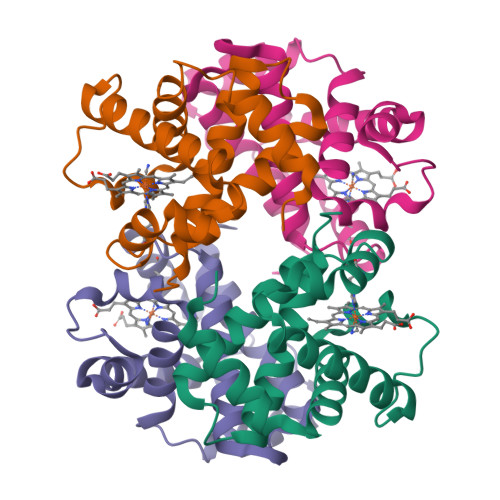
Hemoglobin - an all α helical protein
Close packing in an α-helix
- Helices are very tightly packed structures
- The C=O::HN hydrogen bonds are partially buried within the core of the helical structure
- Because of steric constraints and hydrophobic interactions between side chains, the amino acids have very different propensities for being in an α-helix.
The β-pleated sheet
 |
|---|
| © PDB ID=2QLE |
- Chains are extended
- Interactions are nonlocal
- Can be parallel or antiparallel
- Antiparallel is preferred
- Can contain from 2 to 25+ strands
- Accounts for most non-helical protein structure
- Length: 3.3 Å per residue
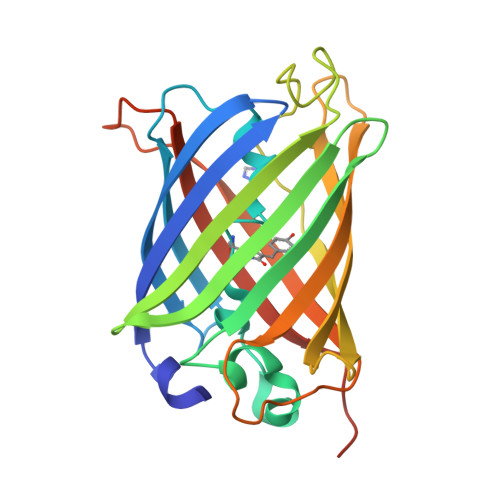
Influenza Neuraminidase an all β-sheet protein
Reverse Turn
- Short 180º turn of ~ 4 residues
- Connects elements of secondary structure
- Interactions are local
- Often occur at surface of protein
- Many types are defined by hydrogen-bonding patterns
The hairpin motif
others
3-10 helix
- Uncommon right-handed helix
- Hydrogen-bonding pattern:
C=0 (i) - - - NH (i+3)- Often found at the ends of α-helices
π helix
- Rare right-handed helix
- Hydrogen-bonding pattern:
C=0 (i)- - - NH (i+5)Ω loop
- Found at the surfaces of proteins
- Base of loop is part of a proteins secondary structure, while the loop is disordered as in the letter omega (Ω )
Random coil
- No regular secondary structure
- Highly flexible
-
Portein
-
Folding collapse;
- Mostly drived by hydrophobic
- hydrogen binding group are fold in the core of the protein
-
Visual
- Schematic; Ribbon diagram; Cα trace; CPK space-filling; Solvent-accessible surface
-
Structure of protein
- Secondary; Tertiary; Quaternary
-
Anfinsen-Merrifield experiments
-
Evolution in sequence
-
protein motif
-
β-sandwich
-
coiled-coil
-
EF hand
-
Domain structure of large proteins
-
Quanternary structure of protein
Protein Folding
Levels of Structure
Exp: Ribonuclease A; 124 residues; 4 disulfide bonds; Unfolded with denaturants (1. urea or guanidine; 2. Oxidase/reduce the disulfide bound.)
Structure motif
- $\beta \alpha \beta$
- $\alpha \alpha \alpha$
- $\beta \beta$
…
Tim-brrel: ($\beta \alpha \beta \alpha$)4
Amino Acid|Graduate Biochemistry 2| Tulane
https://karobben.github.io/2021/08/27/LearnNotes/tulane-biochem-2/









