Nuclear
The central Dogma of Molecular Biology
DAN
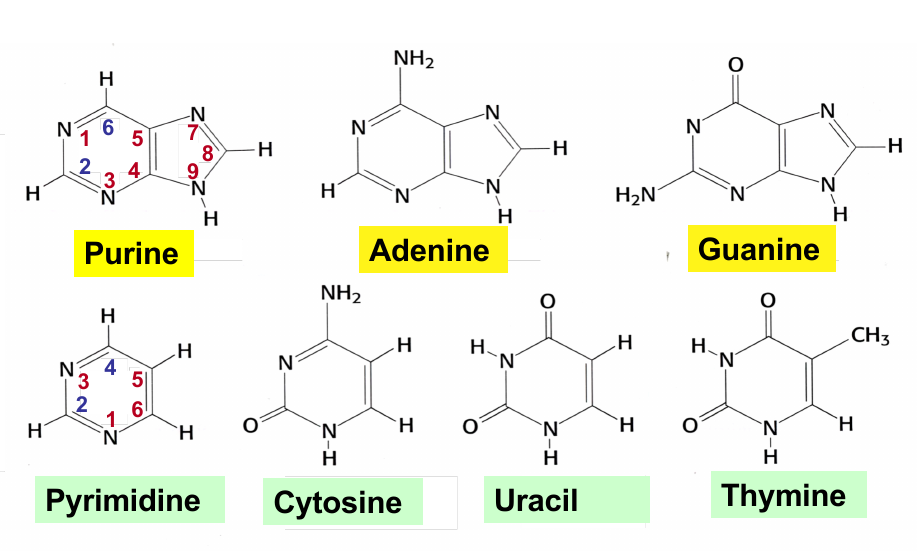
Puring; Pyrimidine
 |
|---|
| © Zac Pursell |
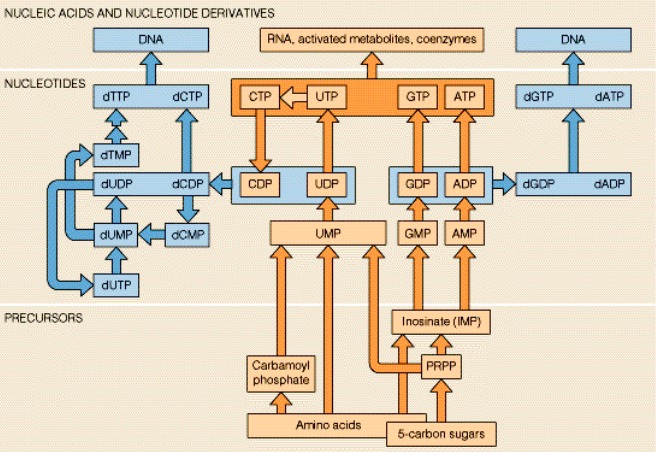
Nucleotides
Biosynthesis of the DNA & RAN purines
Exam: Nucleotide synthesis
OverView
 |
|---|
| © Zac Pursell |
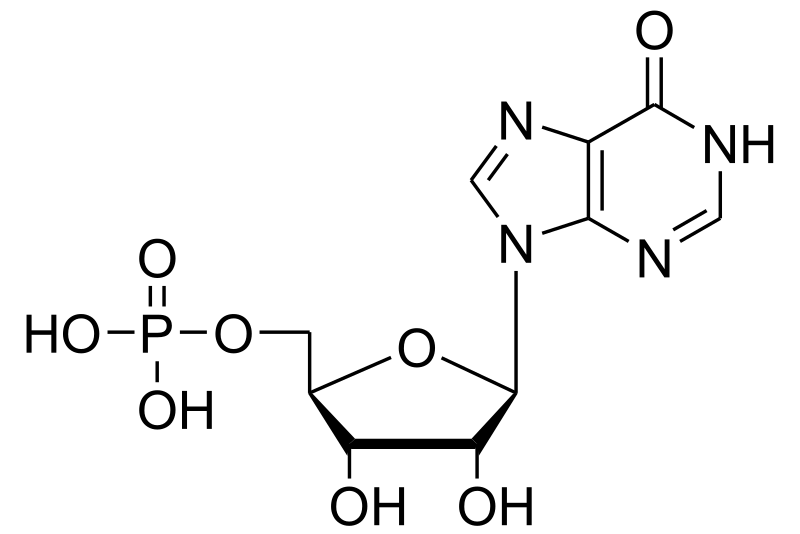
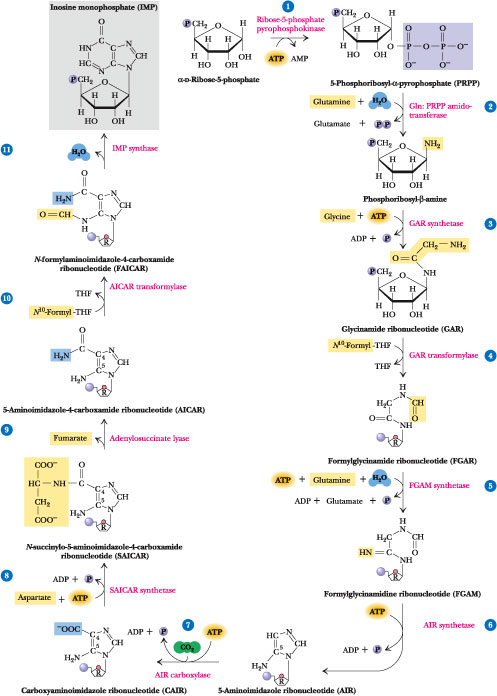
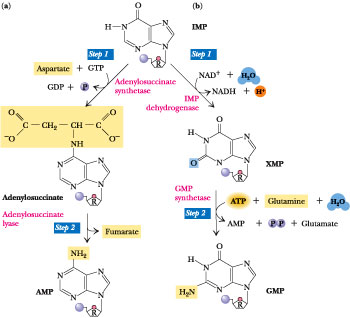
Adenine and guanine nucleotides originate from inosinic acid (IMP), which derives its atoms from phosphoribosyl pyrophosphate (==PRPP=), amino acids, formate and CO2
 |
 |
|---|---|
| © wikipedia | © wikipedia |
Click to see from PRPP to IMP
AMP synthesis requires GTP hydrolysis and aspartate==
GPM syntehsis requries ATP and glutamine
 |
|---|
| © Biochemistry; Garrett & Grisham |
Biosynthesis of the RNA Purine Ribonucleotides
$$
AMP+ATP \overset{NMK}{\leftrightharpoons} 2 ADP + 2NTP \overset{NDK}{\leftrightharpoons} 2ATP + 2NDP
$$
$$
GMP+GTP \overset{NMK}{\leftrightharpoons} 2 GDP + 2NTP \overset{NDK}{\leftrightharpoons} 2GTP + 2NDP
$$
NMK: Nuclioside Monophosphate Kinase
NDK: Nuclioside Diphosphate Kinase
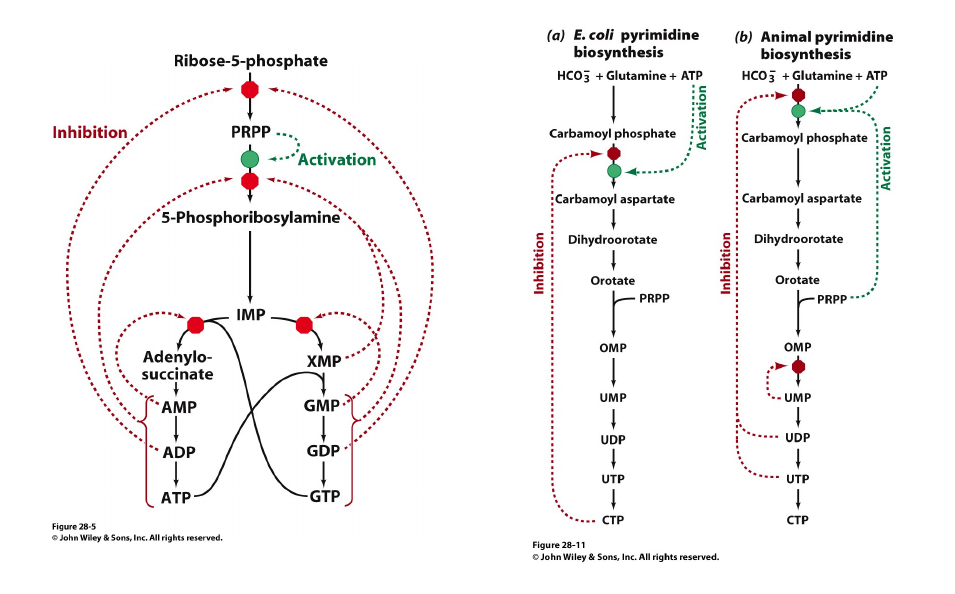
Regulation of the Nucleotide Synthesis
Nucleotide Synthesis is Highly Regulated: Through Feedback Inhibition, Feedforward Activation and Coordinate Regulation
 |
|---|
| © Fundamentals of Biochemistry, 3rd edition |
 |
|---|
| © Biochemistry; Garrett & Grisham |
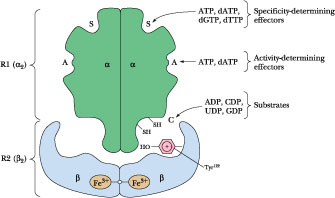
Activity-determining effector (Hexomerization site)
Ribonucleotide reductase is a Allosteric enzyme. Schematic diagram of the quaternary structure. The enzyme consists of two identical pairs of subunits, R12 and R22. Each R2 subunit contains a binuclear Fe(III) complex that generates a phenoxy radical at Tyr 122. The R1 subunits each contain three different allosteric effector sites and five catalytically important Cys residues. The enzyme’s two active sites occur at the interface between the R1 and R2 subunits.
Ribonucleotide Reductases Reduce NDPs to dNDPs
$$
CDP & UDP \underset{dTTP}{\overset{ATP}{\longrightarrow}} dCDP & dUDP
$$
$$
GDP \overset{dTTP}{\longrightarrow} dGDP
$$
$$
NDP \overset{dATP}{\longrightarrow} dNDP
$$
dTMP Is Synthesized from dUMP
Not TDP → dTDP reduction
$UDP \overset{RNR}{\longrightarrow} dUDP \overset{NDP\ kinase}{\longrightarrow} dUTP \overset{dUTPase}{\longrightarrow} dUMP \overset{thymidylate\ synthesis}{\longrightarrow} dTMP → dTTP$
$dUMP + THF \overset{thymidylate\ synthase}{\longrightarrow} dTMP + DHF$
dUTP intermediates are quickly broken down into dUMP to avoid mutagenic dUTP incorporation. The concentration of DHF was quickly rescued by DHFR:
$DHF \overset{DHFR}{\longrightarrow} THF$
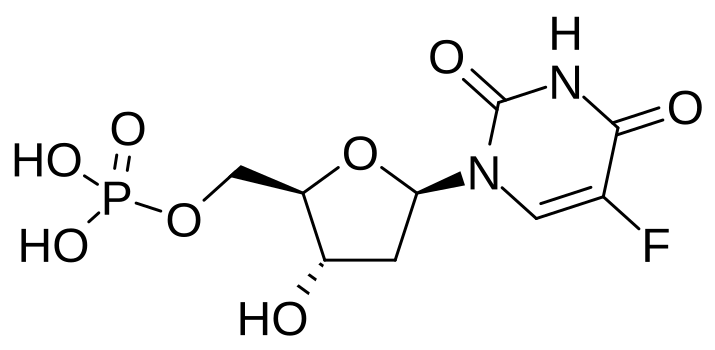
FdU and Methotrexate are anticancer compounds thath disrupt dTMP Biosynthehsis
Cancer require large amount of dTTP than do normal cells.
 |
 |
|---|---|
| © wikipedia | © wikipedia |
| 5-Fluorodeoxyuridylate(FdUMP): locks dTMP synthesis by binding and irreversibly inhibiting thymidylate synthase | Methotrexate: DHF analog; binds tightly and inhibits DHFR |
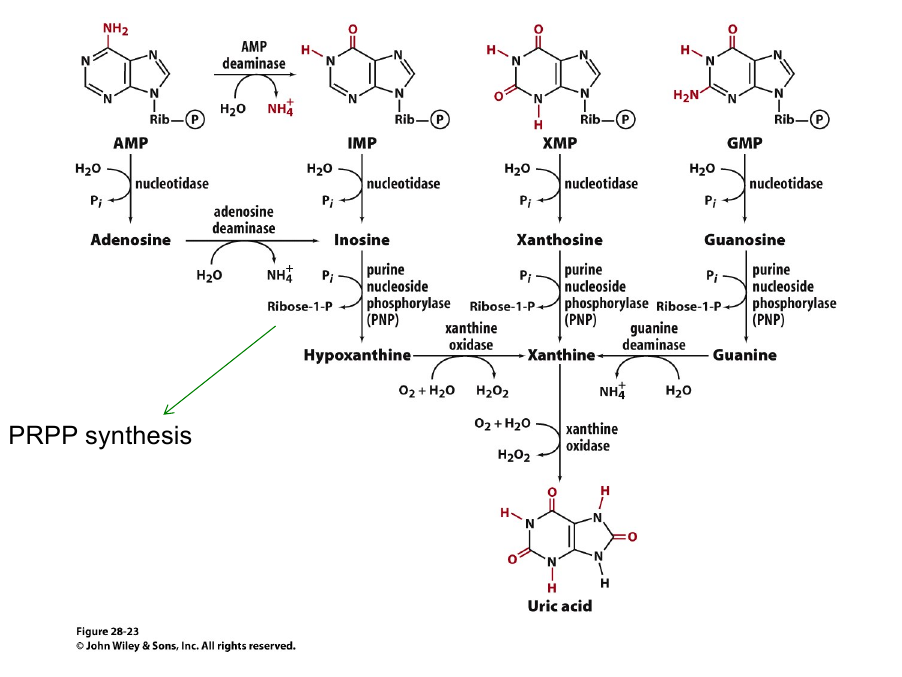
Purines Salvage
Unlike de novo purine syntehsis, purine salvage is NOT conserved across species.
Mammal:
$Adenosine + PRPP \overset{APRT}{\longrightarrow} AMP + PP_i$
$Hypoxanthine/Guanine + PRPP\overset{HPRT}{\longrightarrow} AMP + PP_i$
Animal purin catabolism
 |
|---|
| © Fundamentals of Biochemistry, 3rd edition |
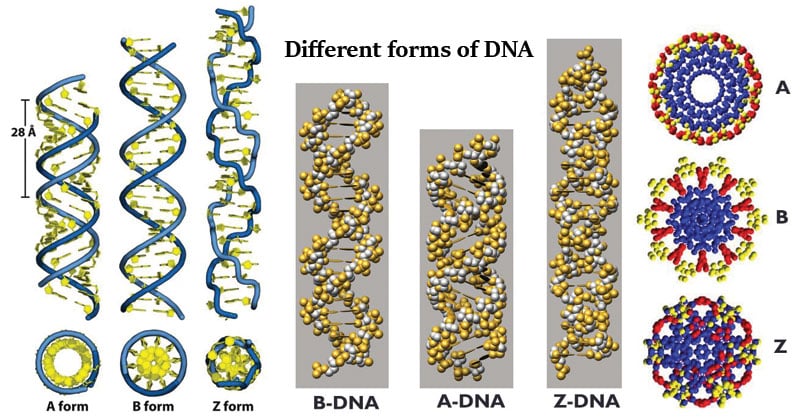
DNA structures and metabolism
The B-DNA Molecule
 |
|---|
| © Sagar Aryal, 2019 |
- Right Handed;
- 20 Å in diameter with two grooves.
- The distance between the bases (rise) is 3.4 Å, 10 bp
- Major groove is ~1.2nm wide, 0.6~0.8nm deep.
- minor groove is ~0.7nm Wide.
B-form by far most common
dsDNA under dehydrating conditions can exist as A-form
Metabolic Instability of RNA
 |
|---|
| © Gail Mitchell Emilsson, et al; 2003 |
RNA is intrinsically unstable in aqueous media and undergoes self-hydrolysis due to nucleophylic attack of the 2 ’OH on the adjacent phosphodiester linkage. This instability increases at alkaline pH.
Base pairing
 |
|---|
| © William Brown |
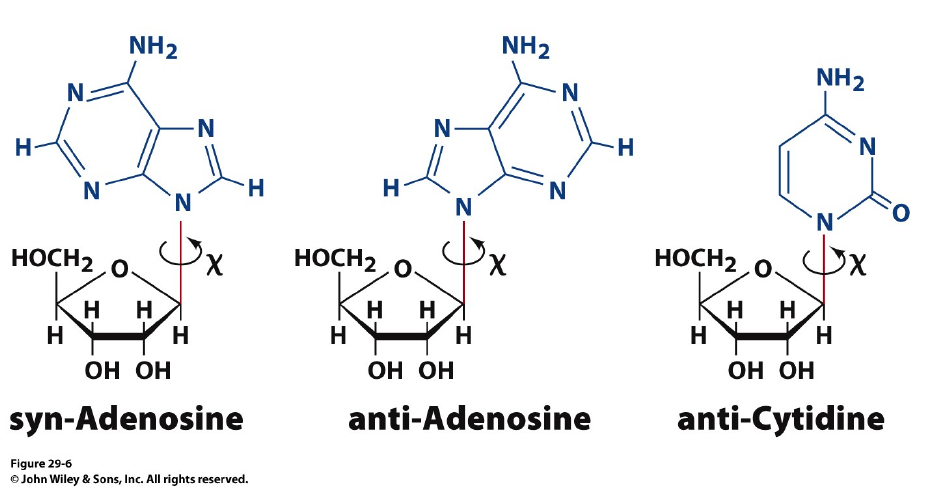
Syn- and anti-configurations: the basis for Z-DNA formation
 |
|---|
| © Fundamentals of Biochemistry, 3rd edition |
Purine nucleotides, particularly guanine nucleotides, have a a tendency to exist in the “syn” configuration. A repeats of GC sequence causes a distortion in the double helix with the phosphates zig-zagging in the backbone.
Sugar-base configurations that affect the conformation of DNA and RNA molecules
 |
|---|
| © wikipedia |
C-2’ endo (South): Distance of phosphates: 7.0 Å; Favored in B-Form DNA
C-3’ endo (North): Distance of phosphates: 5.9 Å; Favored in RNA and some DNA forms
helical conformations
Polynucleotides have a natural tendency to form helical conformations due to stacking interactions between the adjacent hydrophobic bases of the polymer
Hydrophobic forces on planar bases drive stacking interactions
Random coils (unstacked polymers) are favored in alkaline solution, in some organic solvents and at high temperatures
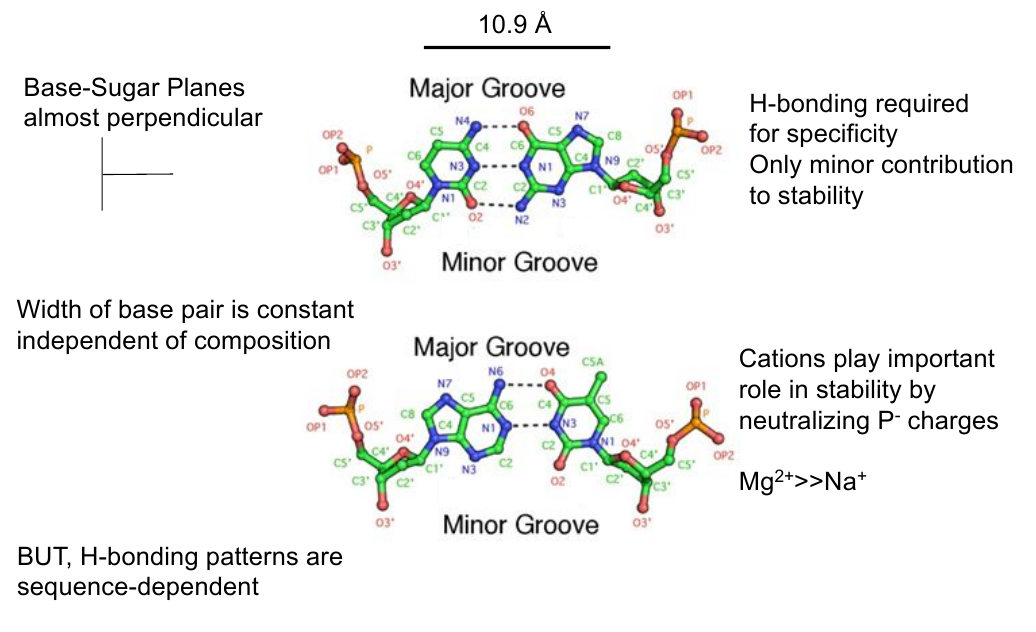
Base Pair Geometry
 |
|---|
| © Zac Pursell |
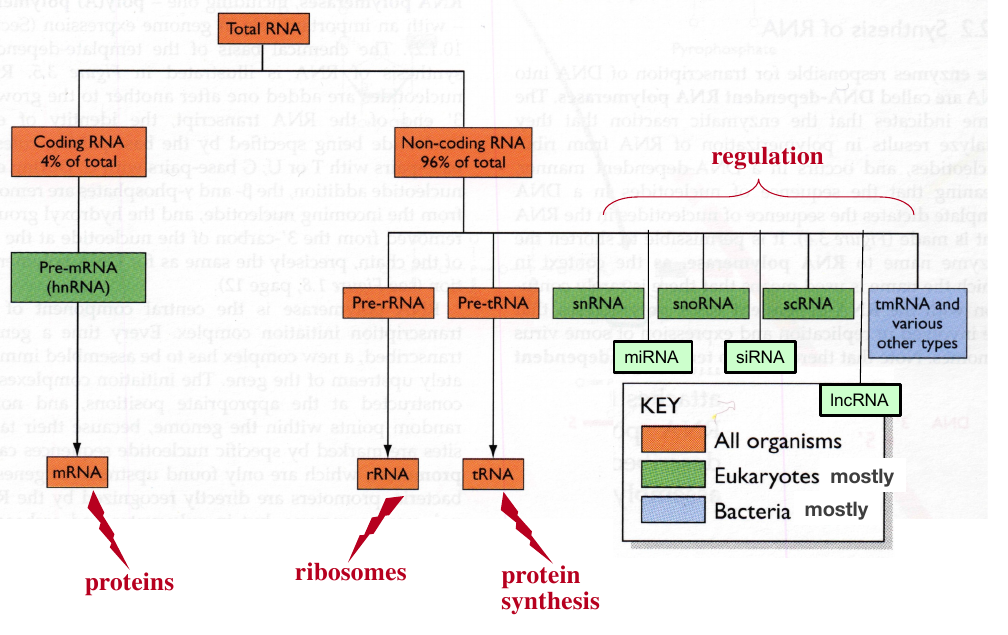
Metabolic roles of cellular RNA
 |
|---|
| © Zac Pursell |
Structure of the RNA
RNA double-helices exist in a variety of shapes
base-pairing, stacking interactions, covalently modified bases, 3 base hydrogen bonding all contribute to compact and complex structure
Ribosomal RNAs
high degree of base pairing
large numbers of “stems” and “loops”
unpaired bases can interact with ribosomal proteins or RNA bases from other RNAs
base-paired segments are mostly α-helical
Some RNAs Capable of Carrying Out Catalysis
self-splicing and self-cleavage reactions in vivo engineered catalysis in vitro
Genomes
- dsDNA, linear, e.g., eukaryotic nuclear DNA, many viruses.
- dsDNA, circular, e.g., bacterial chromosomes, mitochondrial DNA, plasmids, some viruses.
- ssDNA, circular or linear e.g., the genomes of some bacterial, plant and animal viruses.
- ssRNA, e.g., the genomes of many viruses.
- dsRNA, e.g., the genomes of many viruses.
Size of Genome
 |
|---|
| © M. D. Golubovsky, Kenneth Manton; 2005 |








