Wet Lab Techs with Brief Explained
Wet Lab Techs with Brief Explained
Gel Shift Assays–EMSA
What is EMSA
The interaction of proteins with DNA is central to the control of many cellular processes including DNA replication, recombination and repair, transcription, and viral assembly. One important technique for studying gene regulation and determining protein–DNA interactions is the electrophoretic mobility shift assay (EMSA). An advantage of studying protein–DNA interactions by an electrophoretic assay is the ability to resolve complexes of different stoichiometry or conformation. Another major advantage is that the source of the DNA-binding protein may be a crude nuclear or whole cell extract, in vitro transcription product or a purified preparation. EMSA can be used qualitatively to identify sequence-specific, DNA-binding proteins (such as transcription factors) in crude lysates and, in conjunction with mutagenesis, to identify the important binding sequences within the upstream regulatory region of a given gene. EMSA can also be utilized quantitatively to measure thermodynamic and kinetic parameters. @ ThermoFisher 2022
Brief explained: During Gel electrophoration, if we add DNA (oligo) and protein together and the protein could interact with the DNA, the transfer speed of DNA is much slower because protein is dragging them.
One of a standard kit for EMSA:
| LightShift™ Chemiluminescent EMSA Kit from thermofisher. (documentation) |  |
|---|
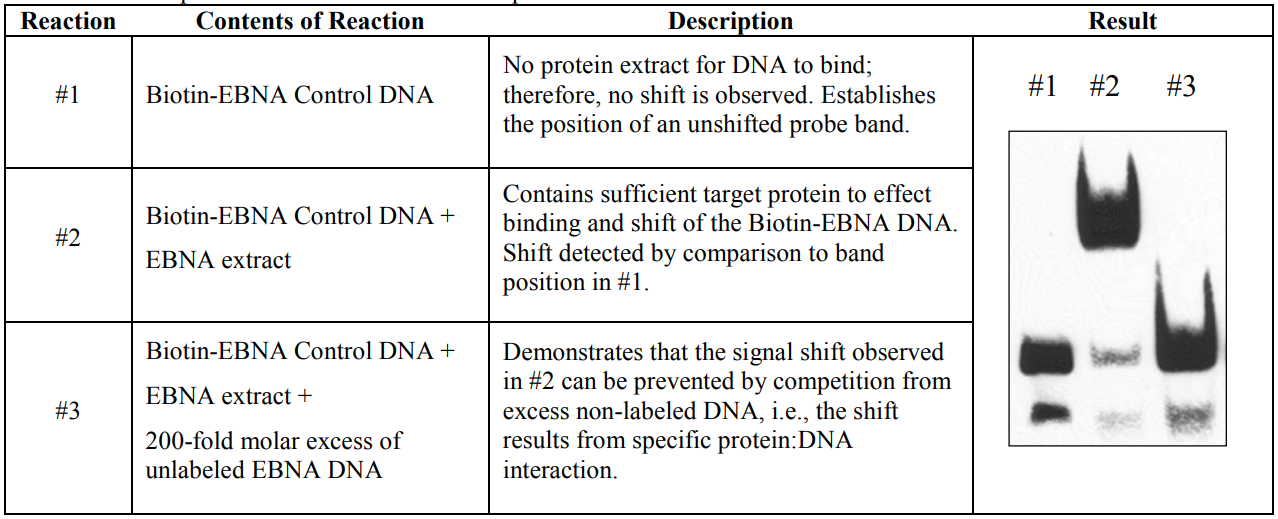 |
|---|
| © ThermoFisher |
According its documentation, a group should have at least 3 tracks:
- labeled DNA: which works as negative control. It shows them band when there are no protein-DNA interaction.
- labeled DNA + protein: The main track for results. The position of DNA shifts because proteins interact with them. The movement of DNA is hindered by proteins.
- labeled DNA + protein + overload unlabeled DNA: By competition, most proteins interact with unlabeled DNA because the quantity of them is higher them labeled. As a result, we can see a dim-shifted band or no shifted band at all. The track could eliminate false positives brought by protein-label tag binding.
Publication used:
- Shyamsunder, Pavithra, et al. “Identification of a novel enhancer of CEBPE essential for granulocytic differentiation.” Blood, The Journal of the American Society of Hematology 133.23 (2019): 2507-2517.
Eamples in paper
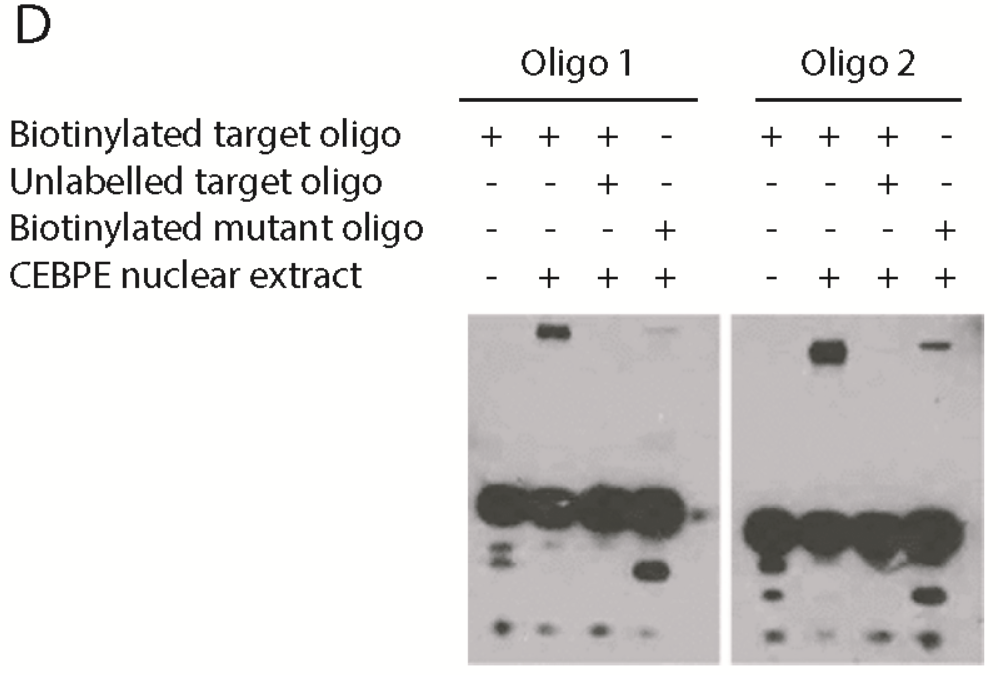 |
|---|
| © Shyamsunder, 2019 |
In this paper, they showed the 4th track, which is protein + unlabeled DNA. In the panel right, there are weak shifted bind which could be the protein.
Luciferase Report Assay
What is a Luciferase Report Assay?
A luciferase reporter assay is a test that investigates whether a protein can activate or repress the expression of a target gene using luciferase as a reporter protein (Carter & Shieh, 2015). The synthesis of the reporter protein and the addition of a substrate results in a chemical reaction with bioluminescence (or the emission of photons) as a by-product. This bioluminescence directly corresponds with the effect of the protein on expression of the target gene. © GoldBio, 2022;
 |
|---|
| © Scholl, et al., 2014 |
 |
| © GoldBio, 2022; |
Scholl, Zackary & Yang, Weitao & Marszalek, Piotr. (2014). Chaperones Rescue Luciferase Folding by Separating Its Domains. The Journal of biological chemistry. 290. 10.1074/jbc.M114.582049.
After inserting the target DNA segment in the plasmid, the vector is transferred into cells. If proteins could interact with the segment, luciferase would express and cells could illuminate.
 |
|---|
| © Chrysostomos Tornari; 2010 |
Wet Lab Techs with Brief Explained








