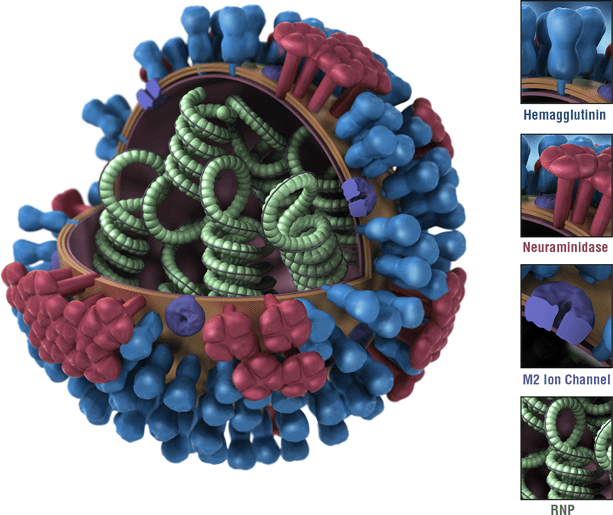
Neuraminidase (NA) protein, a Quick View
NA protein
The influenza NA protein, or neuraminidase, is a critical component of the influenza virus and plays a vital role in the virus’s life cycle. Here’s an overview of its function and structure:
Function of Influenza NA Protein
-
Viral Release: Neuraminidase is primarily responsible for facilitating the release of newly formed virus particles from the host cell. It cleaves sialic acid residues on the host cell surface and on the viral envelope, which otherwise bind the emerging viral particles and prevent their release.
-
Virus Spread: By cleaving sialic acid, NA protein aids in the spread of the virus. This cleavage prevents the aggregation of virus particles, enabling them to spread more efficiently from cell to cell.
-
Role in Infection: It helps the virus penetrate the mucus layer of the respiratory tract, enhancing the virus’s ability to infect the host cells.
Structure of Influenza NA Protein
 |
|---|
| © Julie L. McAuley1; 19 |
-
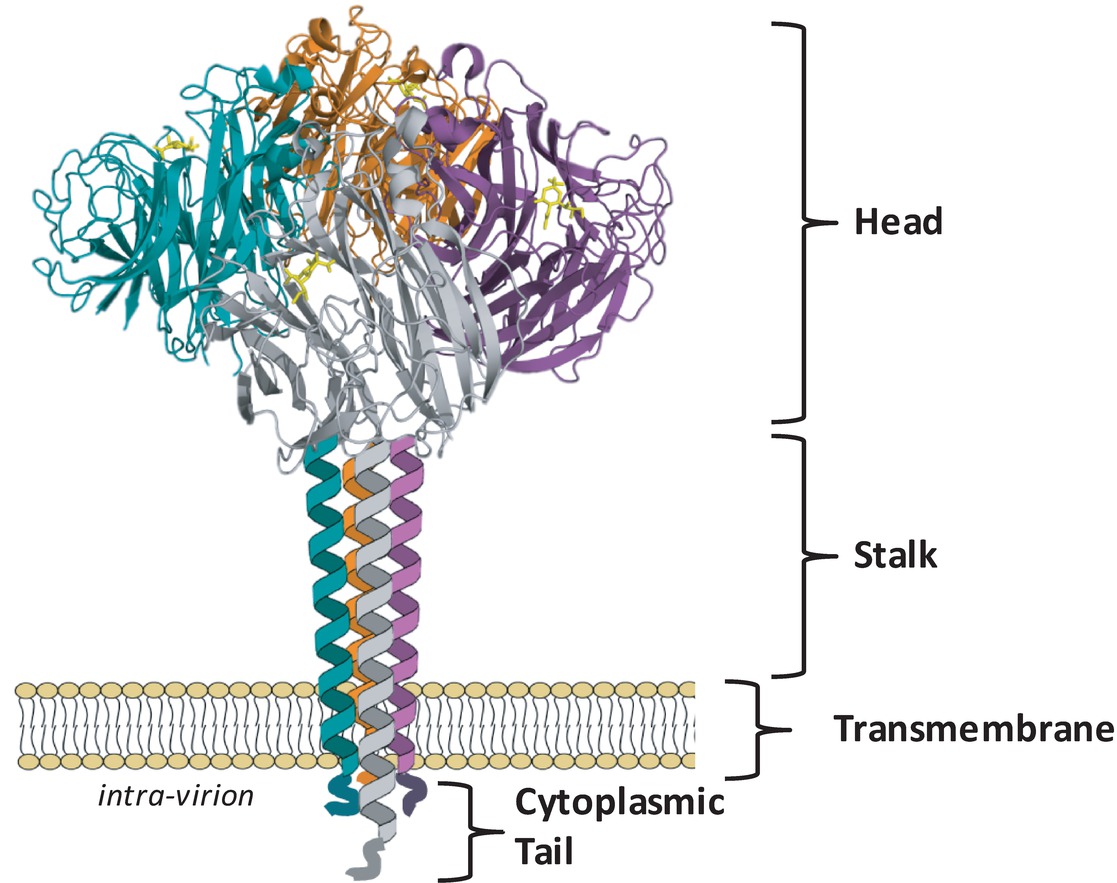
Cytoplasmic Tail: Suggesting that the NA cytoplasmic tail is involved in critical viral functions, the N-terminal domain sequence is nearly 100% conserved across all IAV subtypes and consists of the sequence MNPNQK[1]. A complete loss of the tail domain[2] resulted in a 50% reduction in the amount of NA in infected cells.
-
Transmembrane Domain: It contains a variable sequence of amino acids spanning residue numbers 7–29 and is predicted to form an alpha helix[1:1][3] with interspersed polar residues driving subunit-subunit interactions[4].
-
Stalk Region: Depending on the length of the stalk region, the NA may protrude slightly more or less above the viral envelope than the HA, which may influence the overall enzymatic activity of the virus[5][6]. The number and sequence of amino acid residues can vary considerably[1:2]. NA stalk truncation mutants of the 2009 pandemic virus A(H1N1)pdm09 showed greater lethality in mice and virulence in ferrets than the untruncated counterpart[7].
-
Head Domain: These catalytic sites are characterized by a large cavity with an unusually large number of charged residues in the pocket and around its rim[8][9]. The tetrameric form of NA is considered optimal for enzyme activity, and mutations that lead to instability of the tetramer lead to decreased enzyme activity [McKimm_96b][Fujisaki_12][^McKimm_13]. While it has been reported that monomers alone have no enzyme activity[3:1] and usually expression of recombinant soluble NA heads requires a synthetic tetramerization domain for active NA[10].
- Active site: Arg118, Asp151, Arg152, Arg224, Glu276, Arg292, Arg371, and Tyr406
- Framework residues: Glu119, Arg156, Trp178, Ser179, Asp198, Ile222, Glu227, Glu277, Asn294, and Glu425
-
Tetrameric Structure: The neuraminidase protein typically forms a tetramer, meaning four NA molecules join together to function.
-
Head and Stalk Regions: The NA protein has two distinct regions: the head, which contains the active site for sialic acid cleavage, and the stalk, which anchors the protein to the viral envelope.
-
Active Site: The active site is located in a pocket in the head region and is responsible for the enzyme’s sialic acid cleavage activity.
-
Glycosylation Sites: These sites are present on the head region, where carbohydrate chains are attached. This glycosylation can affect the antigenicity and activity of the NA protein.
-
Subtypes: There are several different subtypes of neuraminidase, categorized based on slight variations in their amino acid sequences. These variations can affect the protein’s function and its recognition by the immune system.
Understanding the function and structure of the influenza NA protein is crucial for developing antiviral drugs and vaccines. Neuraminidase inhibitors, for example, are a class of antiviral drugs that block the activity of this protein, effectively preventing the virus from spreading within the host.
Leading Research
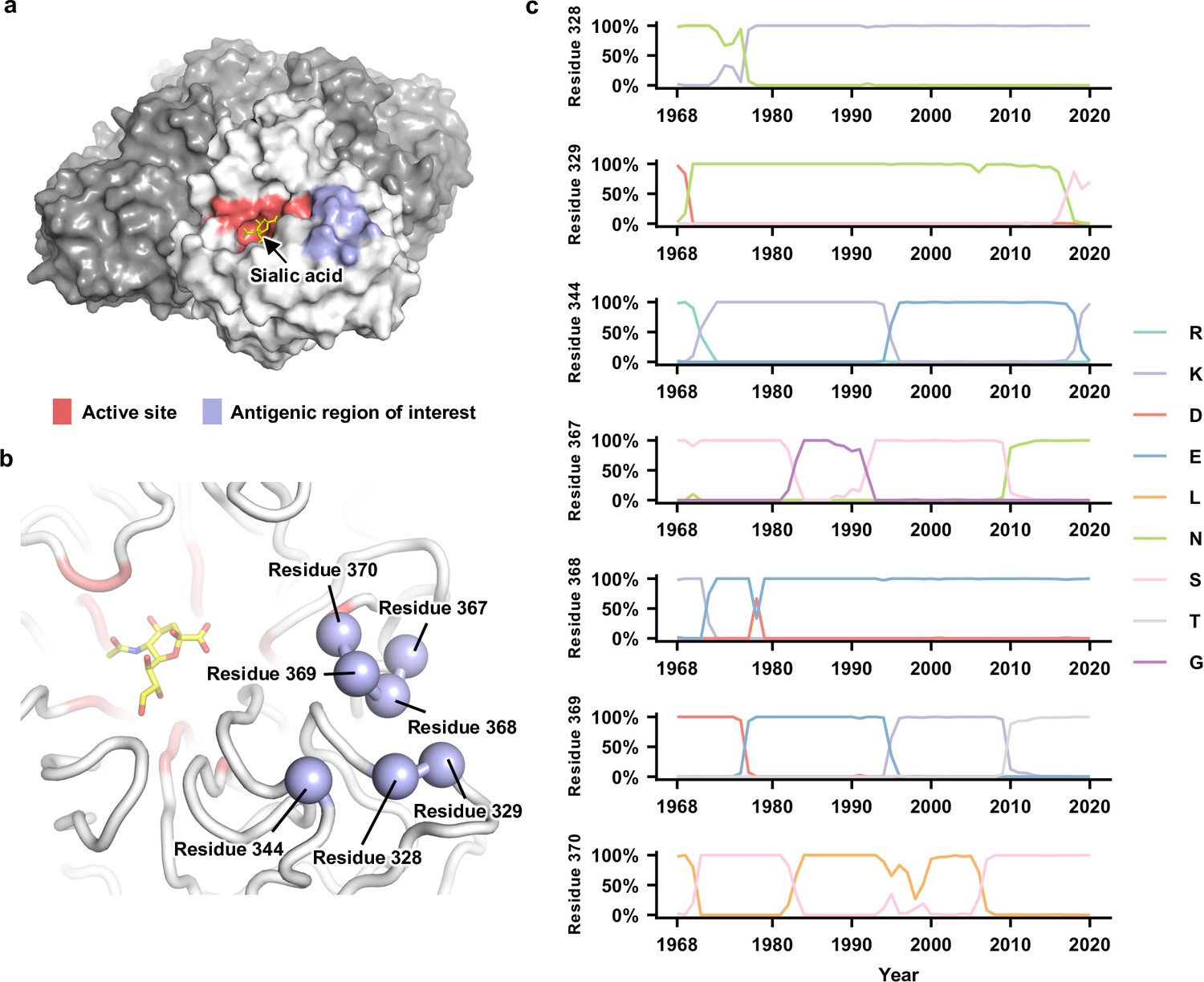
A paper[11] investigates the antigenic evolution of the neuraminidase (NA) protein in the H3N2 influenza virus. Using combinatorial mutagenesis and next-generation sequencing, the study focuses on seven specific residues in an antigenic region of NA. It finds that the local fitness landscape of this region is highly correlated across different H3N2 strains and reveals that local net charge balancing is a significant constraint in NA antigenic evolution. The study also demonstrates a correlation between epistasis and residue coevolution in naturally circulating influenza strains, providing important insights into the biophysical constraints on NA antigenic evolution.
 |
|---|
| © Wang Y; 2021[11:1] |
Features from Other NA protein
H1N1:
- K253R could reduce the 75% of virion-associated NA activity[12].
Blok, J., and Air, G. M. (1982). Variation in the membrane-insertion and “stalk” sequences in eight subtypes of influenza type A virus neuraminidase. Biochemistry 21, 4001–4007. ↩︎ ↩︎ ↩︎
Garcia-Sastre, A., and Palese, P. (1995). The cytoplasmic tail of the neuraminidase protein of influenza A virus does not play an important role in the packaging of this protein into viral envelopes. Virus Res. 37, 37–47. doi: 10.1016/0168-1702(95)00017-K ↩︎
Air, G. M. (2012). Influenza neuraminidase. Influenza Other Respir. Viruses 6, 245–256. doi: 10.1111/j.1750-2659.2011.00304.x ↩︎ ↩︎
Nordholm, J., Da Silva, D. V., Damjanovic, J., Dou, D., and Daniels, R. (2013). Polar residues and their positional context dictate the transmembrane domain interactions of influenza A neuraminidases. J. Biol. Chem. 288, 10652–10660. doi: 10.1074/jbc.M112.440230 ↩︎
Harris, A., Cardone, G., Winkler, D. C., Heymann, J. B., Brecher, M., White, J. M., et al. (2006). Influenza virus pleiomorphy characterized by cryoelectron tomography. Proc. Natl. Acad. Sci. U. S. A. 103, 19123–19127. doi: 10.1073/pnas.0607614103 ↩︎
Matsuoka, Y., Swayne, D. E., Thomas, C., Rameix-Welti, M. A., Naffakh, N., Warnes, C., et al. (2009). Neuraminidase stalk length and additional glycosylation of the hemagglutinin influence the virulence of influenza H5N1 viruses for mice. J. Virol. 83, 4704–4708. doi: 10.1128/JVI.01987-08 ↩︎
Park, S., Il Kim, J., Lee, I., Bae, J. Y., Yoo, K., Nam, M., et al. (2017). Adaptive mutations of neuraminidase stalk truncation and deglycosylation confer enhanced pathogenicity of influenza A viruses. Sci. Rep. 7:10928. doi: 10.1038/s41598-017-11348-0 ↩︎
Colman, P. M., Varghese, J. N., and Laver, W. G. (1983). Structure of the catalytic and antigenic sites in influenza virus neuraminidase. Nature 303, 41–44. doi: 10.1038/303041a0 ↩︎
Varghese, J. N., McKimm-Breschkin, J. L., Caldwell, J. B., Kortt, A. A., and Colman, P. M. (1992). The structure of the complex between influenza virus neuraminidase and sialic acid, the viral receptor. Proteins 14, 327–332. doi: 10.1002/prot.340140302 ↩︎
Schmidt, P. M., Attwood, R. M., Mohr, P. G., Barrett, S. A., and McKimm-Breschkin, J. L. (2011). A generic system for the expression and purification of soluble and stable influenza neuraminidase. PLoS One 6:e16284. doi: 10.1371/journal.pone.0016284
[^McKimm_13]McKimm-Breschkin, J. L., Williams, J., Barrett, S., Jachno, K., McDonald, M., Mohr, P. G., et al. (2013). Reduced susceptibility to all neuraminidase inhibitors of influenza H1N1 viruses with haemagglutinin mutations and mutations in non-conserved residues of the neuraminidase. J. Antimicrob. Chemother. 68, 2210–2221. doi: 10.1093/jac/dkt205
[^Fujisaki_12]Fujisaki, S., Takashita, E., Yokoyama, M., Taniwaki, T., Xu, H., Kishida, N., et al. (2012). A single E105K mutation far from the active site of influenza B virus neuraminidase contributes to reduced susceptibility to multiple neuraminidase-inhibitor drugs. Biochem. Biophys. Res. Commun. 429, 51–56. doi: 10.1016/j.bbrc.2012.10.095
[^McKimm_96b]McKimm-Breschkin, J. L., McDonald, M., Blick, T. J., and Colman, P. M. (1996b). Mutation in the influenza virus neuraminidase gene resulting in decreased sensitivity to the neuraminidase inhibitor 4-guanidino-Neu5Ac2en leads to instability of the enzyme. Virology 225, 240–242. ↩︎Wang Y, Lei R, Nourmohammad A, et al. Antigenic evolution of human influenza H3N2 neuraminidase is constrained by charge balancing[J]. Elife, 2021, 10: e72516. ↩︎ ↩︎
Liu T, Wang Y, Tan T J C, et al. The evolutionary potential of influenza A virus hemagglutinin is highly constrained by epistatic interactions with neuraminidase[J]. Cell Host & Microbe, 2022, 30(10): 1363-1369. e4. ↩︎
Neuraminidase (NA) protein, a Quick View