Hemagglutinin, the Influenza Virus Protein
 © PDB |
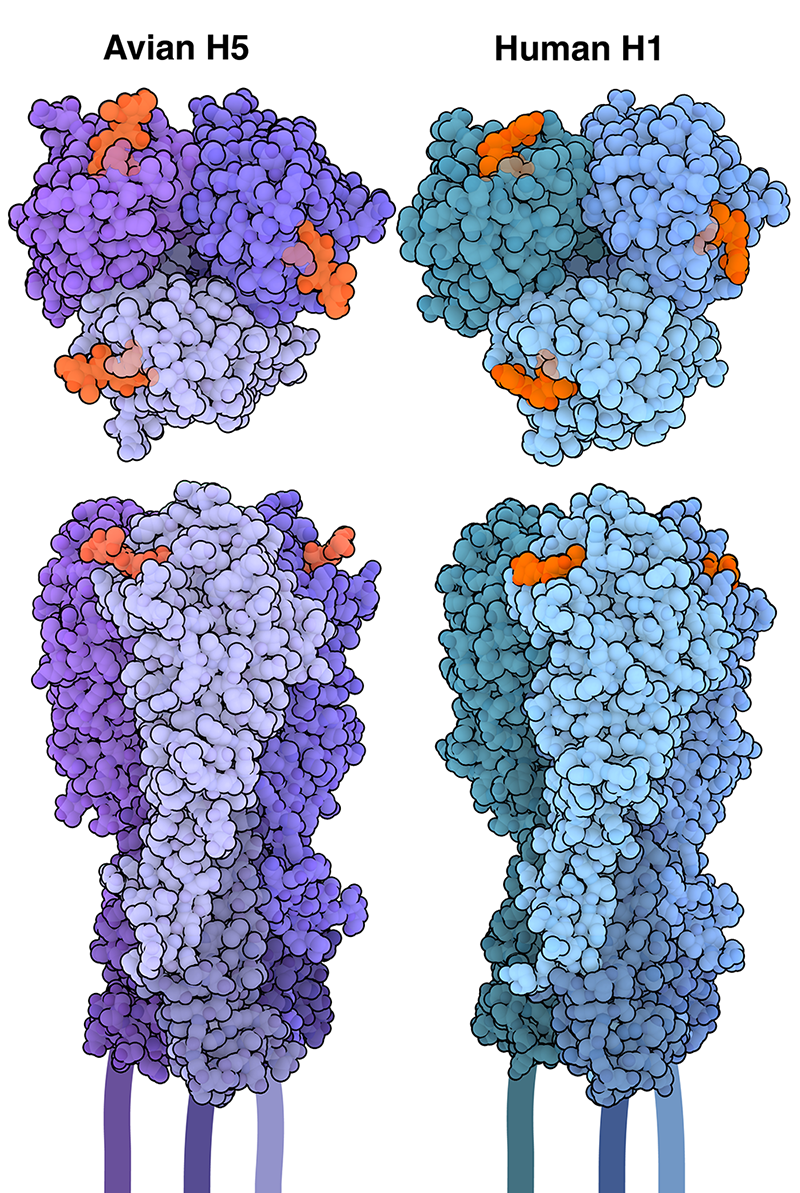
Influenza is able to enter and infect cells through the action of hemagglutinin, which recognizes and attaches to specific molecules on the cell surface. Most hemagglutinins have been found to target sialic acids, a family of nine-carbon sugar molecules that are commonly found at the tips of glycans, or sugar chains, that are attached to proteins and lipids on the cell surface. Sialic acid can be linked to glycans in different ways. |
 © Donald J. Benton, 2018 |
(A) Cryotomogram section showing cross-section of a liposome with examples of HAs tilted with respect to the lipid bilayer (white boxes). (Scale bar: 20 nm.) (B) Gallery of subtomograms of tilted HA in liposomes. Images in the second row are identical to those above but indicate HAs (blue) and liposome bilayer (red lines). (Scale bar: 10 nm.) |
Sialic Acid
 |
 |
|---|---|
| Birds sialic acid (Neu5Ac) with α-2,3 bound whith also happen in human deep lung | α-2,6 sialic acid (Neu5Ac) bound with also happen in human upper respiratory tract. |
© Dr.G Bhanu[1]
Hemagglutinin Cleavage
 © Dr.G Bhanu[1:1] |
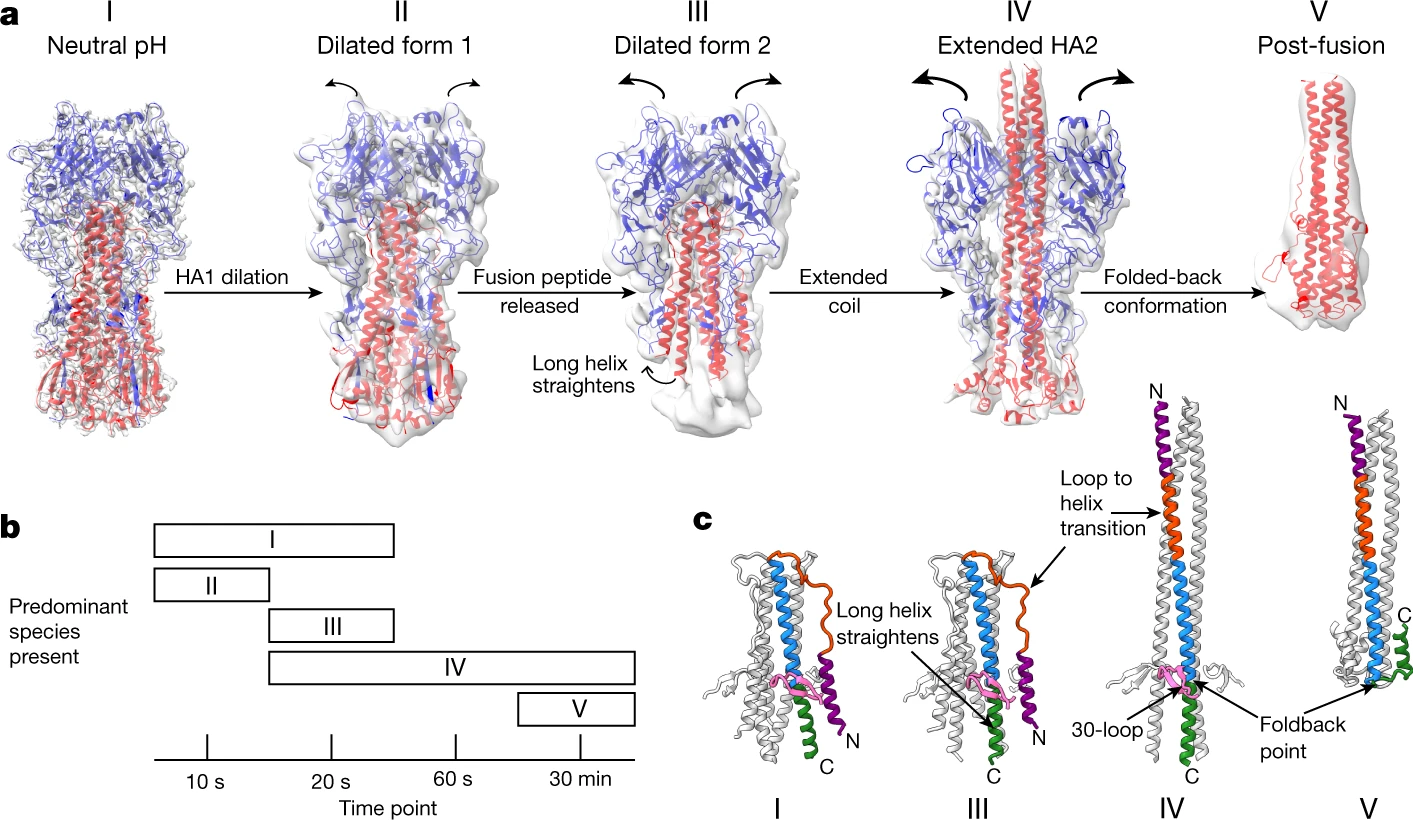
Hemagglutinin (HA) has lots of conformations. The HA0 is the original form of the protein on the virus surface. It then cleaved into HA1 and HA2 by host proteases in the endosomes. Proteases only cleave inner cell HA0[2]. After the cleavage, the fusion peptid is exposed which is in the end of the HA2 |
Conformation Change of Hemagglutinin
 |
 |
|---|
At low pH in endosomes, about pH 5.5, depending on the virus strain, fusion of virus and endosomal membranes is activated in a process that involves extensive changes in HA conformation[3]
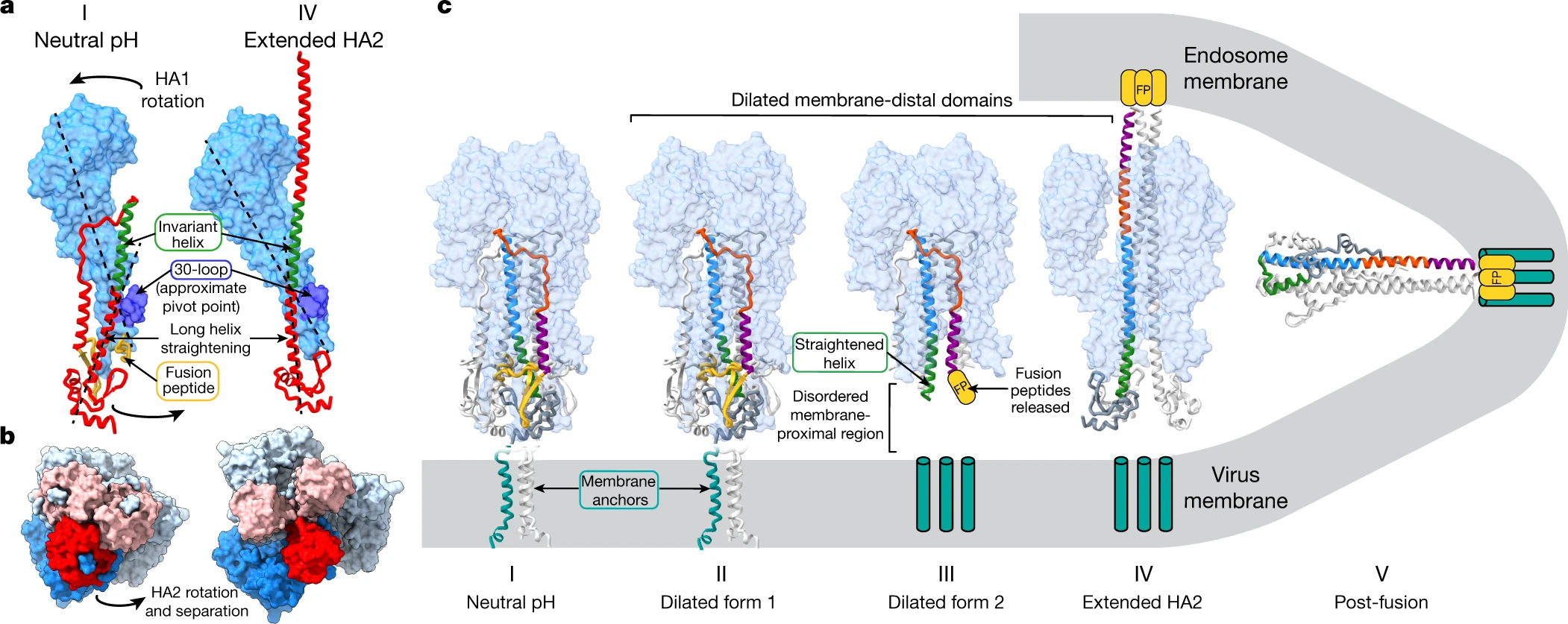
Anchor of the HA Protein
Donald J. Benton, et al., observe the structure of the region that anchors HA in the virus membrane as a bundle of three α-helices that are joined to the ectodomain by flexible linkages. And the fab, HA−FISW84, binds to near there (6HJQ) my affects the flexibility of the linker and prevents the conformational changes that are necessary for membrane fusion[4].
 |
The structure of the membrane-associated region. Detailed views of (A) tilted and (B) straight micelles, as shown in Fig. 1 B and C respectively. |
|---|
© Donald J. Benton, 2018: The flexible linker region (purple) runs between Gly-175, at the C terminus of the 160 helix, and Gly-182 and extends to the N termini of the α-helices of a trimeric α-helical bundle, residues 186 to 203 (cyan). (C) The amino acid sequence of the transmembrane domain. The sequence shown begins at the 160 helix to the C terminus of HA2. Color-coded block diagrams indicate the positions of these structural elements in the sequence.
Fusion Model
 |
A 3.2-nm-thick computational slice through a reconstructed cryo-electron tomogram shows several examples of HA bridging (blue arrowheads) between virus particles and 80% DOPC:20% Chol liposomes after 30 s of incubation at pH 5.5[5] |
 |
|---|
 |
| © Lee lab |
Zhirnov O P, Ikizler M R, Wright P F. Cleavage of influenza a virus hemagglutinin in human respiratory epithelium is cell associated and sensitive to exogenous antiproteases[J]. Journal of virology, 2002, 76(17): 8682-8689. ↩︎
PA Bullough, FM Hughson, JJ Skehel, DC Wiley, Structure of influenza haemagglutinin at the pH of membrane fusion. Nature 371, 37–43 (1994). ↩︎
Benton D J, Nans A, Calder L J, et al. Influenza hemagglutinin membrane anchor[J]. Proceedings of the National Academy of Sciences, 2018, 115(40): 10112-10117. ↩︎
Gui L, Ebner J L, Mileant A, et al. Visualization and sequencing of membrane remodeling leading to influenza virus fusion[J]. Journal of virology, 2016, 90(15): 6948-6962. ↩︎
Hemagglutinin, the Influenza Virus Protein
https://karobben.github.io/2025/03/13/LearnNotes/hemagglutinin/









