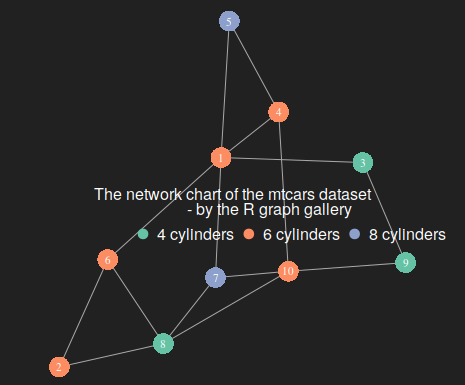
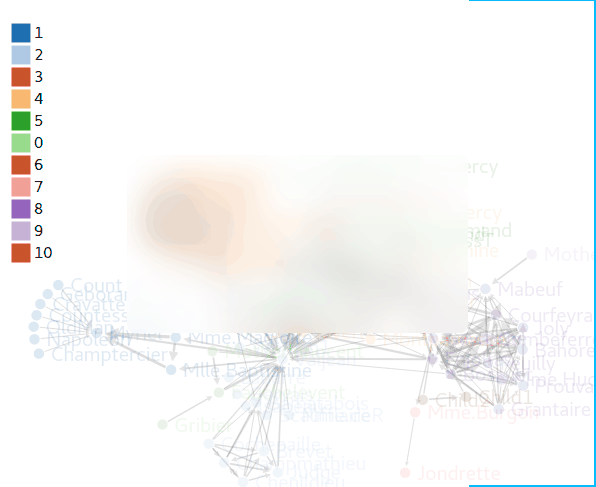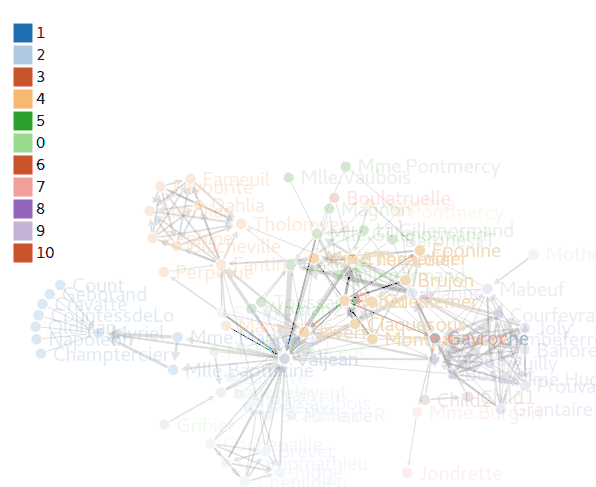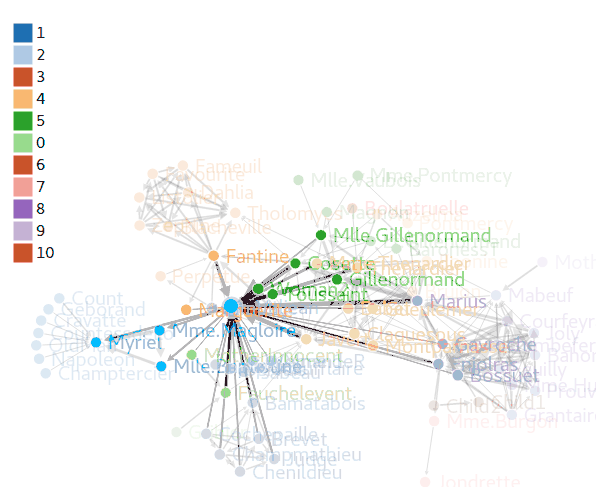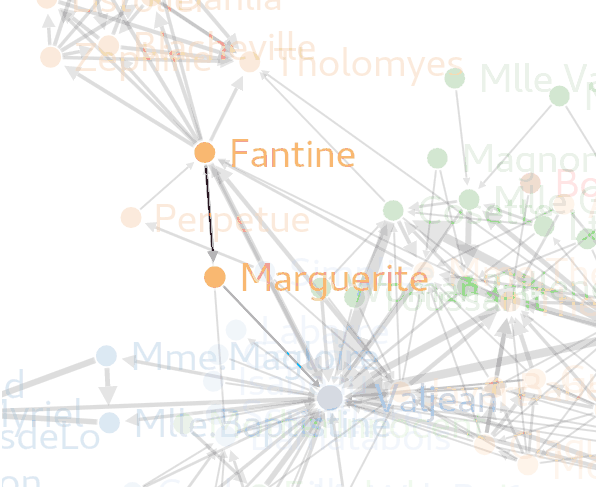
forceNetwork(Links=genelinks,
Nodes=genenodes,
Source="source",
coul = brewer.pal(nlevels(as.factor(mtcars$cyl)), "Set2")
指定Links文件中的源节点
Target="target",
linkColour=genelinks$col,
Value="value",
NodeID="name",
fontSize=20,
Group="group",
opacity=0.8,
zoom=TRUE,
arrows=TRUE,
opacityNoHover=0.7,
legend=TRUE,
height=600,
width=600
)
MyClickScript <- 'r = confirm(d.name+"\\n\\nIf you\'d like to know more about "+d.name+", Please Click \'OK\'");
if (r == true){
window.open("https://www.uniprot.org/uniprot/"+d.name+"_HUMAN");};'
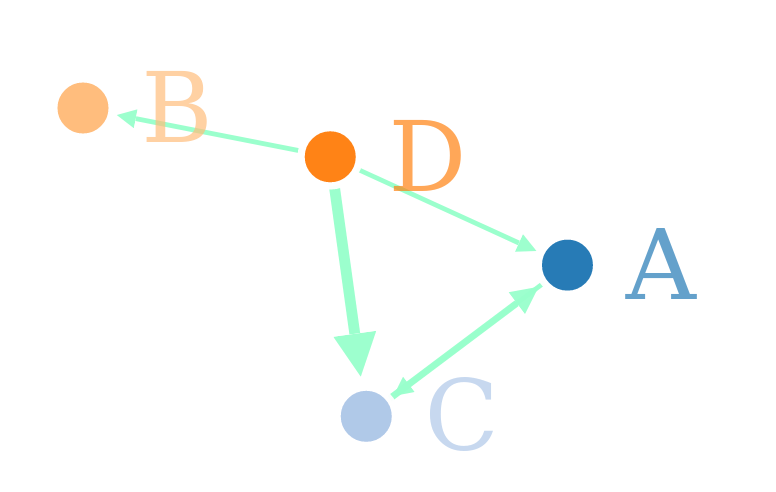
forceNetwork(
Links=GeneLinks,
Nodes=GeneNodes,
Source="source",
Target="target",
linkColour="#99FFCC",
Value="value",
NodeID="name",
Nodesize = "Freq_l",
fontSize=20,
Group="group",
opacity=0.98,
zoom=TRUE,
arrows=F,
opacityNoHover=0.7,
legend=TRUE,
clickAction = MyClickScript)
|