Protein Analysis|Graduate Biochemistry 6| Tulane
Purification and Analysis Overview
- Methods of protein separation and purification
- Protein solubility
- Isoelectric focusing
- Principles of chromatography
- Polyacrylamide gel electrophoresis
- Gel filtration
- Ion exchange/Hydrophobic interaction
- Affinity Chromatography
- Ultracentrifugation
Methods of protein analysis- Summary of primary information
- Protein sequencing by Edman degradation
- Mass spectrometry
- Methods of determining protein three dimensional structure
Methods of Macromolecule Separation
| Method of Separation | Physical Basis | sensitivity | Specifity |
|---|---|---|---|
| Precipitation | Solubility (charge, isoelectric point) | μg - kg | Very low |
| Mass Spectrometry | Molecular Mass | fg - ug | Very High |
| Nondenaturing PAGE | Charge/mass, shape | μg - mg | High |
| SDS PAGE | Molecular Weight | μg - mg | High |
| Isoelectric focusing | Isoelectric point | μg - mg | Moderate |
| Capillary Electrophoresis | Charge/mass | ng - μg | High |
| Gel Filtration | Size and shape | ng - mg | High |
| Ion Exchange | Net charge | ng - mg | Moderate High |
| Hydrophobic interaction Chromatography | Hydrophobicity | ng - mg | High |
| Affinity Chromatography | Specific molecular interactoin | ng - mg | Very High |
| Density Gradient Ultracentrifugation | Molecular weight, density and shape | μg - g | Moderate |
| Equlibrium Density Ultracentrifugation | Density | μg - g | Moderate |
Legend:
- Electrophoresis techniques
- Chromatography techniques
- Ultracentrifugation techniques
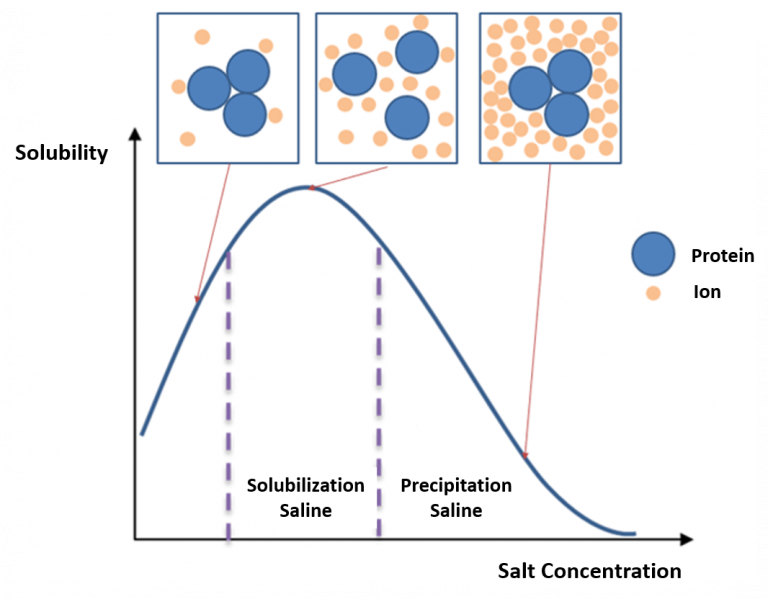
Protein Solubility in Salt
Salting in
- Increase in solubility with increasing salt at low ionic strength
- Proteins unfold at low ionic strength and irreversibly aggregate
Salting out
- Protein precipitation at high ionic strength driven by ionic charge shielding and solvent effects
- Solubility is minimum at isoelectric point (ionic effect)
- Effectiveness of salting out in Hoffmeister series (solvent effect):
- Anions: SO4>H2PO4>CH3COO>Br>I>ClO4>SCN
- Cations: NH4,Cs,K,Na>Li>Mg>Ca>Ba
PS: Increase the salt, you shield the charge surface of the protein
 |
 |
|---|---|
| © Michel Awkal 2020 | © Richard R Burgess 2009 |
Solting in: The unsaturated ions on the surface of the protein can interacted with other ions to increase the soluability of the protein.
Isoelectric focusing
 |
|---|
| © Blake C Meyers 2007 |
Isoelectric point (pI) is the pH at which positive and negative charges are equally abundant on a molecule.
pI for the G is ~6;
pI for D is ~3.5
pI for K is ~ 9.5
- Isoelectric focusing is equilibrium electrophoresis
- Ampholytes - Mixtures of buffer molecules covering a range of pKa values stabilize the pH gradient in an electric field
- Charged molecules, including the ampholytes, will move toward the location where their net charge is zero. This is the isoelectric point, or pI.
Chromatography
Principle of Chromatography
- Molecules in a mobile medium move through a stationary phase with which they can interact.
- Differential interactions with the stationary phase are the basis for physical separation of molecules
Some stationary phases
- Paper (paper Chromatography)
- Bare silica (Thin layer chromatography)
- Functionalized Silica (Column Chromatography)
- Various polymers (acrylamide, dextran, agarose, and others)
Mobile phase is usually liquid
SDS PAGE
Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis
Acrylamide gel:
- Polymerized acrylamide
- Macromolecule movement is hindered
- Movement driven by electric field
- Mobility is determined by charge, size and shape
SDS: A detergent that denatures proteins
- with/withough reducing agent:
- breaking the disulfide bond
Proteins denatured in SDS:
- 3 SDS bound per amino acid
- Constant charge/mass
- Mobility α ln(mass)
- SDS can also solubilize hydrophobic AA
- Useful but may lots some information.
Gel Filtration Chromatography
 |
 |
|---|---|
| © Marcell Wolf, 2015 | © Rufika Shari Abidin |
Vo = Void Volume, Excluded Volume
Vi = Included Volume
VT (Total solvent volume) = Vo+ViFor each mocromolecule
Vp = Penetrable volume
Ve (Protein Elution Volume) = Vo+Vpσ (fractional retention) = Vp/Vi
Vp/Vi = (Ve - Vo)/(VT - Vo)
- Polymer pours beets.
- separated the protein by its size
- You can’t tell the protein is monomor, dimer or etc.
- small proteins randomly filtered inside of the beets and causing transfer delay.
Ion Exchange Chromatography
Retention is due to ionic interactions with fixed charges on stationary phase
Counterions compete for binding sites
Three phases: binding, elution, regeneration
Binding:
- Cation exchange: fixed anions
- Anion exchange: fixed cations
Typical Ionic groups
- Diaminoethylamino (DEAE)
- Carboxymethyl (caboxylate)
- Sulfoethyl/Sulfobenzyl (sulfate)
The material is charged:
- Same charge: not interacted
- opposite Charge: Reacted
Hydrophobic Interaction Chromatography
- Binding to stationary phase is hydrophobic
- Elution is with organic solvents (Acetonitrile, methanol, isopropanol, hexane)
- Often used in High Pressure Liquid Chromatography (HPLC).
Affinity Chromatography
Retention is due to specific interaction
Typical affinity molecules:
- Antibody
- Ligand/substrate/inhibitor
- Lectin (carbohydrates)
- Nickel-Nitrilotriacetate or Ni-NTA Binds to engineered poly Histidine
PS: Some specific interact between the protein and the ?agent
Example of Enzyme Purification
| Purification Step | Total Acitivity | % recovery | Specific Activity |
|---|---|---|---|
| Crude Extract | 31800 | 100 | 1 |
| Acid Precipitate | 59900 | 189 | 10.1 |
| Amm. Sulfate Precipitate | 57200 | 175 | 27.2 |
| GTP ( affinity) | 37300 | 117 | 3962 |
| Mono Q (ion exchange) | 14300 | 45 | 6304 |
| Gel filtration | 11000 | 35 | 11620 |
Ultracentrifugation
Bouyant force depends on:
- Density Difference between molecule and solvent
Frictional force depends on:
- Molecular shape
- Solvent viscosity
Centrifugal force depends on:
- Molecular weight
- Rotation speed
- Distance from center
Ultracentrifugation
- Can be used to obtain information on molecular weight, molecular shape and intermolecular interactions
- Sedimentation velocity (S=Svedbergs)
- Sedimentation equilibrium
Density Gradient Ultracentrifugation
- Sucrose gradient:
- Gradient is man-made and impermanent
- Sample applied at top
- Separation is by sedimentation velocity
CsCl density Gradient:
- Gradient is formed by centrifugal force
- Equilibrium position of macromolecule determined by its density
Protein Sequence Determination
Primary Information
- Number of chains
- Dansyl Chloride reaction (Voet, Voet & Pratt pg 109)
- One cycle of Edman degradation
- SDS PAGE (under reducing conditions)
- Primary Sequence
- Fragmentation, purification of fragments and Edman sequencing or mass spectroscopy
- Requires overlapping fragments
- Common fragmentation methods:
- Cyanogen Bromide (cleaves C-term side of Met)
- Trypsin (cleaves C-term side of Arg, Lys)
- Chymotrypsin (cleaves C-term side of Trp, Phe, Tyr)
- Disulfide linkages
- SDS PAGE (reduced and non-reduced)
- Fragmentation, purification and partial sequencing with disulfide bonds oxidized/reduced
Protein Sequencing
Principles of Edman degradation:
- Cyclic, sequential removal and identification of N-terminal amino acids
- Identification is done by High Pressure Liquid Chromatography (HPLC)
- Can sequence 50-100 residues
- Sequence determination requires overlapping fragments
- Derivitization: Phenylisothiocyanate (PITC)
- Acid cleavage: PTC polypeptide terminal remove
- Acid reduction: Polypeptide like of residue
- Separation and Identification
(Back to 1.)
cite: Voet, Voet & Pratt pg 113
Mass Spectrometry
Principle of Mass Spectrometry
- Acceleration of a molecule in an electric field depends on the mass/charge ratio
Protein Mass Spectrometry
MALDI TOF - Matrix assisted laser desorption - Time of flight
ESI - Electrospray ionizationBoth methods can be used to determine the molecular weight of proteins up to ~ 105 Daltons
Molecular weight determination is unambiguous
Proteomics - by mass spectrometry
Genomics: The study of the an organism’s genetic information.
Proteomics: The study of large scale protein expression pattern in a cell type or tissue.Genomic information is fixed. Proteomic information depends on life cycle, environment, cell type etc.
Below: Two-dimensional gel electrophoresis of an E. coli whole cell extract.
Modern MALDI/SELDI mass spectrometers can identify these proteins
Often done by cutting out spots, digesting proteins with trypsin and getting the mass of the fragments by mass spec.
Determination of Protein Structure
- Two methods are used to determine protein structure:
- X-ray crystallography
- Two-dimensional Nuclear Magnetic Resonance (NMR) spectroscopy
X-ray crystallography
- purify protein
- grow crystals
- collect X-ray diffraction patterns
- determine electron density in three dimensions
- solve protein structure
Advantages:
- Can solve very large proteins/complexes
- Gives atomic-level resolution
Disadvantages:
- crystallization is often difficult or impossible
- crystal contacts can distort structure
Determination of Protein Structure
2D NMR Spectroscopy
- purify protein
- collect multidimensional NMR spectra in solution
- assign peaks and crosspeaks to specific residues
- calculate distances between residues from the strength of the crosspeaks
- solve structure using distance constraints
Advantages:- no crystallization required
- structure is a solution structure
Disadvantages:- Only small proteins (<200 residues) can be solved
- Resolution is not at atomic level
Protein Analysis|Graduate Biochemistry 6| Tulane
https://karobben.github.io/2021/09/24/LearnNotes/tulane-biochem-6/









