Notes for Genetics
Who knows = =
Extra DNA, why?
transposon
DAN transposon:
- cut & paste
Retrotransposon: - copy & paste
DNA transposon:
transpoase recognize the IR sequences and cut it out.
Form a Pre-integration complex
Jump into another site of DNA
IR: inverted repeat
TSD: target site duplication (direct repeat)
 |
|---|
| exp: © The Tn3-family of Replicative Transposons |
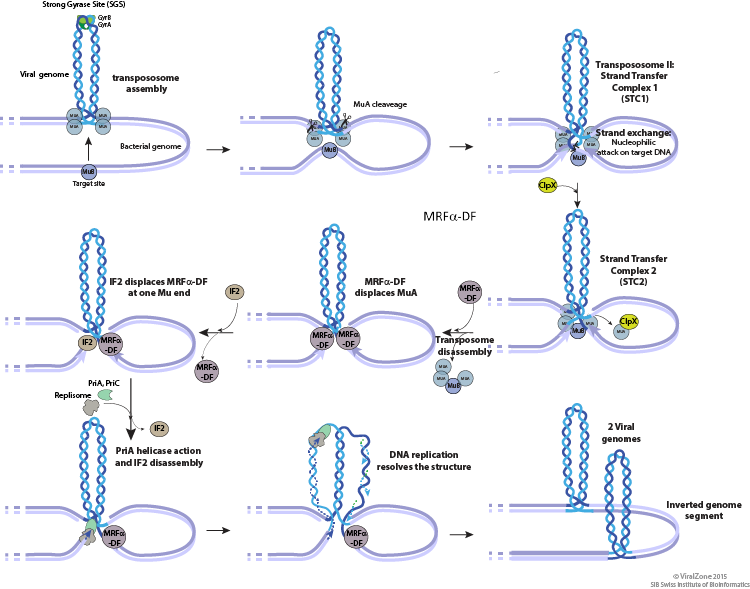 |
|---|
| © viralzone |
Retrotransposons
Retrotransposons have shared similarity with retroviruses
Reverse transcriptase
Similar in life cycle, similar in gene structure.
Consequence of transpositon
- Insertion of a sequence in a new site
- Cutting of DNA strands
Generally: mutagenic processes, generally bad in excess
Potential disadvantages:
- disruption of coding sequence
- deletion/rearrangements of the sequences
- non-allelic homologous recombination (based on similar seq contributed by transposons)
- alternative splicing
- Epigenetic alterations
- “transduction” of neighboring sequnces
- premature polyadenylation or other transcription defects
Benefits:
V(D)J recombination in vertebrates
- V(D)J recombination generates diversity in immune system
- The RAGI.RAG2 complex catalyses the programmed deletions during antibody rearrangement.
Exp a dramatic example of active retrotransposition that is “put to use” by the hose - Drophila telomeres
non-LTR retrotransposons replaces the function of the telomerase to extending the end of the linear chromosomes.
P elements, more information need
P elements are transposable elements that were discovered in Drosophila as the causative agents of genetic traits called hybrid dysgenesis. The transposon is responsible for the P trait of the P element and it is found only in wild flies. They are also found in many other eukaryotes.[1]
Generate Mouse models
- Spontaeous mutant starins
- chemical-induced mutatgenesis models
- Transplatntation/Engraftment models
- Transgenic and gene targeting models
- PDX mouse models.
PS: PDX can preserve the traits better than other for some reason.
Spontaneously arising mutatnt strain lethal yellow
- Maturity-onset obesity
- Adipocytehypertrophy
- hypertriglyceridemia
- Hyperinsulinemia
- Diabetes
- Hypatic lipogenesis
- Increased caloric efficiency
Chmical mutagensis mouse models
- Germline: N-ethyl-N-nitrosourea ENU (1/700 loci) (cause random mutations in sperm)
- Pancreas: Streptozotocin (destroys pancreatic β-cells), T1D
- Skin: 7,12-dimethylbenz[a]anthracene (DMBA) + TAP
- Lung: Nitrosamines
- Linve
- Bearst;
- Colon:
- Bladder
Cheap, quick, and generate lots of mutatios
- Hard to figure out where, when, and how it cause the mutation.
ENU-induced mutatgenesis generates random mutat strains
- Gene knockout → gene1 → Phenotype 1, 2, 3
- Genome-wide mutatgenesis → lots of lots of genes → Lots of interested phynotypes
Forward and reverse Genetic Screens
-
Forward:
-
Spontaneously arising
-
Phynotype Screen
-
Mutation phenotypes reveal gene functions
-
Map the genes and identify the gen products
-
Reverse genetic screen
ENU - induced mutagenesis
it oxidize the G and destroy the hydrogen bond of the G
Forward genetic screen using ENU-induced mutagenesis
Patient-dereived xenograft (PDX) Model
Plant cancers from human to growth on mouse.
HGPS
- HGPS
- Muscular dystrophy
- Cardiomyopathy
- Lipodystrophy
- Mutation from laminopathies, Laminin A
Laminin A is a structural protein that forms nuclear lamina in inner nuclear membrane nuclear lamina is importatnt for maintatining interphase nuclear architecture heterozygous C to T substitution at 1824, G608G.
Reducing LMNA gene dosage rescues Zmpste24 KO-induced progera syndrome phnetype
Sketch for CH12
Target:
- How Regulated
- Variaty ways for regulation
- Chromatin in regulation
- Epigemetic Markers
- RNA regulation (miRNA, post-TGR)
- Difference between the eukaryotic cells and prokaryotic cells:
- One RNA pol in prokaryotic but 3 in Eu
- Intron and exon
Majumdar, S; Rio, DC (April 2015). “P Transposable Elements in Drosophila and other Eukaryotic Organisms”. Microbiology Spectrum. 3 (2): MDNA3–0004–2014. doi:10.1128/microbiolspec.MDNA3-0004-2014. PMC 4399808. PMID 26104714. ↩︎
Notes for Genetics
https://karobben.github.io/2022/01/24/LearnNotes/tulane-genetics-1/








