CD-HIT
Documentation: CD-HIT
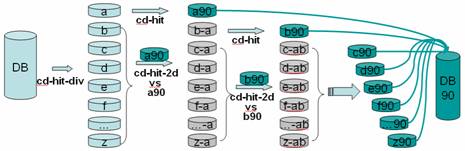
CD-HIT (Cluster Database at High Identity with Tolerance) is a widely used bioinformatics tool designed to cluster biological sequences (such as DNA, RNA, or proteins) to reduce sequence redundancy and improve the efficiency of other sequence analyses. It’s particularly useful when dealing with large datasets, such as those frequently encountered in genomics and proteomics studies.
Here are some key features and uses of CD-HIT:
-
Sequence Clustering: CD-HIT efficiently clusters similar sequences together based on a user-defined similarity threshold. For example, if you set a threshold of 90%, the tool will group sequences that are 90% identical into the same cluster.
-
Reduction of Redundancy: By clustering similar sequences, CD-HIT helps in reducing redundancy in the dataset. This is particularly important when creating sequence databases or when analyzing large datasets where many sequences may be very similar or nearly identical.
-
Speed and Efficiency: CD-HIT is known for its speed and low memory usage, making it suitable for handling very large datasets that are common in modern high-throughput sequencing projects.
-
Multiple Applications: It’s used in a variety of applications, such as creating non-redundant sequence databases, preparing datasets for further analyses (like phylogenetic studies), or in metagenomics for analyzing diversity and similarity of species.
-
Customizable Parameters: Users can customize several parameters, such as the identity threshold for clustering, the word size for initial comparisons, and memory usage, allowing for flexibility depending on the specific requirements of the data or the analysis.
-
Output Files: CD-HIT generates two main types of output files - one containing the clustered sequences and another (
.clstrfile) detailing the composition of each cluster.
CD-HIT is a command-line tool, which means it is run from a terminal or command prompt and offers great flexibility in scripting and automation within bioinformatics pipelines. The tool is an essential part of the toolkit for biologists and bioinformaticians dealing with large-scale sequence data.
Example
|
The command you’ve provided is for CD-HIT, a widely used bioinformatics tool for clustering and comparing protein or nucleotide sequences. CD-HIT helps to significantly reduce the redundancy of large datasets by clustering similar sequences together based on a specified sequence identity threshold. This is particularly useful in tasks like constructing databases or preparing datasets for other analyses where redundancy might be an issue.
Let’s break down the components of your command:
-
cd-hit: This invokes the CD-HIT program. -
-i Trunked.fa: The-ioption specifies the input file. Here, the input file isTrunked.fa, which likely contains a collection of nucleotide or protein sequences in FASTA format. -
-o cdhit/Trunked_$x.fa: The-ooption specifies the output file. This command will create an output file in thecdhitdirectory with a name based on the value of the variable$x. The$xseems to be a shell variable that would be replaced with its value when the command runs. -
-c $x: The-coption sets the sequence identity threshold. Sequences are clustered together if they are similar to each other above this threshold. The exact value is determined by the variable$x. -
-M 32000: This sets the maximum available memory (in MB).32000here means 32,000 MB, or 32 GB of RAM. -
-d 0: The-doption is for the description length to show in the.clstroutput file.0means it will show the full sequence defline. -
-T 8: This specifies the number of threads to use.8means the program will run on 8 threads, enabling parallel computation for faster processing. -
-n 5: For protein sequences, this sets the word length for the initial match.5is typically used for thresholds between 0.7 and 0.8. -
-aL 0.8: This sets the alignment coverage for the longer sequence.0.8means 80% of the longer sequence must be covered in the alignment. -
-s 0.95: This sets the alignment coverage for the shorter sequence.0.95means 95% of the shorter sequence must be included in the alignment. -
-uS 0.2: This specifies the maximum unmatched percentage of the shorter sequence.0.2means up to 20% of the shorter sequence can be unmatched. -
-sc 1: This option, when set to1, sorts clusters by decreasing size. -
-sf 1: This option, when set to1, sorts fasta sequence by cluster size.
Each of these options tailors the behavior of CD-HIT to your specific requirements, influencing how sequences are clustered and how the output is formatted and organized. The use of shell variables (like $x) suggests that this command is part of a script where the value of $x is dynamically set, allowing for flexible application of the command with different parameters.
Read the cd-hit Result as pandas dataframe
|
Trouble Shooting
Fatal Error: Too low cluster threshold for the word length. Increase the threshold or the tolerance, or decrease the word length. Program halted !!
The error message you’re encountering, “Fatal Error: Too low cluster threshold for the word length. Increase the threshold or the tolerance, or decrease the word length. Program halted !!”, indicates a conflict between the parameters you’ve set in CD-HIT, particularly relating to the sequence identity threshold and the word length.
In CD-HIT, the “word length” refers to the length of the initial exact match that the algorithm looks for when comparing sequences. The “cluster threshold” is the percentage similarity required for two sequences to be clustered together. When the cluster threshold is set too low relative to the word length, the program cannot effectively perform the initial sequence matching, leading to this error.
To resolve this issue, you can:
-
Increase the Threshold: Increase the sequence identity threshold (
-cparameter). If it’s currently very low, increasing it will make the clustering less stringent. For instance, if you have set-c 0.6(60% identity), try increasing it to 0.7 or higher. -
Decrease the Word Length: Decrease the word length (
-nparameter). A lower word length means the program will look for shorter exact matches in the initial step, which can be more tolerant of low similarity thresholds. However, be cautious, as too low a word length might make the process slower and less efficient. -
Adjust Tolerance: If your dataset includes very diverse sequences and you need to maintain a low threshold, you might have to adjust other parameters to balance the sensitivity and specificity of the clustering.
Remember that the appropriate values for these parameters can vary depending on your specific dataset and the objectives of your analysis. It might require some experimentation to find the right balance for your needs.