How to do anova in R
Anova test, all you need to know and action in R
Citation:
- ANOVA Test: Definition, Types, Examples, SPSS; © Statistics How To
- ANOVA in R | A Complete Step-by-Step Guide with Examples; © scribbr
In this post, I’ll show how to do ANOVA test in R. The result would be similar like Prim. If it is not, I’ll noted below.
To be noticed, we should carefully select which protocol we use based on our data and purpose. For example, one-way ANOVA, two-way ANOVA, pairwise comparing based on the TukeyHSD test, control group pairwise comparing based on Dunnett Test, and finally, the repeat ANOVA test. As far as I know, we do not have a way to do ANOVA based on the summarized data set. However, we can simulate a table based on the summarized data and do the ANOVA test which could achieve the same result as the Prism, cheers!
Talk is cheap, show me the code
raw posts: Rebecca Bevans: ANOVA in R | A Complete Step-by-Step Guide with Examples
Example data: Click to Download
|
density block fertilizer yield
1:48 1:24 1:32 Min. :175.4
2:48 2:24 2:32 1st Qu.:176.5
3:24 3:32 Median :177.1
4:24 Mean :177.0
3rd Qu.:177.4
Max. :179.1
|
One way anova
|
summary(one.way)
Df Sum Sq Mean Sq F value Pr(>F)
Time 1 2042344 2042344 1349 <2e-16 ***
Residuals 576 872212 1514
Tukey multiple comparisons of means
95% family-wise confidence level
Fit: aov(formula = weight ~ Diet, data = ChickWeight)
$Diet
diff lwr upr p adj
2-1 19.971212 -0.2998092 40.24223 0.0552271
3-1 40.304545 20.0335241 60.57557 0.0000025
4-1 32.617257 12.2353820 52.99913 0.0002501
3-2 20.333333 -2.7268370 43.39350 0.1058474
4-2 12.646045 -10.5116315 35.80372 0.4954239
4-3 -7.687288 -30.8449649 15.47039 0.8277810
Tow way anova
|
Df Sum Sq Mean Sq F value Pr(>F) Diet 3 155863 51954 11.504 2.57e-07 *** Chick 46 374243 8136 1.802 0.00136 ** Residuals 528 2384450 4516 Tukey multiple comparisons of means 95% family-wise confidence level Fit: aov(formula = weight ~ Diet * Chick, data = ChickWeight) $Diet diff lwr upr p adj 2-1 19.971212 0.3164389 39.62599 0.0447928 3-1 40.304545 20.6497722 59.95932 0.0000011 4-1 32.617257 12.8550000 52.37951 0.0001458 3-2 20.333333 -2.0257975 42.69246 0.0896286 4-2 12.646045 -9.8076279 35.09972 0.4678041 4-3 -7.687288 -30.1409612 14.76638 0.8140383 $Chick diff lwr upr p adj 16-18 12.71428571 -203.752079 229.180650 1.0000000 15-18 23.12500000 -190.313714 236.563714 1.0000000 13-18 30.83333333 -175.368054 237.034721 1.0000000 9-18 44.16666667 -162.034721 250.368054 1.0000000 20-18 41.41666667 -164.784721 247.618054 1.0000000 10-18 46.08333333 -160.118054 252.284721 1.0000000 8-18 55.00000000 -152.536038 262.536038 1.0000000 17-18 55.50000000 -150.701387 261.701387 1.0000000
Anova with Mean and sd table
Reference: Dason, 2015
The best way to calculate the Anova table from the summary data in R is not to do the ANOVA directly, is to simulate a data table based on your summary data and run ANOVA after.
|
Df Sum Sq Mean Sq F value Pr(>F) group 5 16310 3262 3.85 0.00256 ** Residuals 157 133011 847 Tukey multiple comparisons of means 95% family-wise confidence level Fit: aov(formula = y ~ group, data = simulated_data) $group diff lwr upr p adj 2-1 -19 -43.409541 5.409541 0.2230739 3-1 -1 -25.775796 23.775796 0.9999970 4-1 9 -16.676208 34.676208 0.9135955 5-1 -13 -37.409541 11.409541 0.6412141 6-1 -17 -40.540078 6.540078 0.3012203 3-2 18 -4.458911 40.458911 0.1952944 4-2 28 4.551540 51.448460 0.0093845 5-2 6 -16.054213 28.054213 0.9697289 6-2 2 -19.087861 23.087861 0.9997892 4-3 10 -13.829491 33.829491 0.8310731 5-3 -12 -34.458911 10.458911 0.6379762 6-3 -16 -37.510748 5.510748 0.2694050 5-4 -22 -45.448460 1.448460 0.0795394 6-4 -26 -48.541958 -3.458042 0.0136716 6-5 -4 -25.087861 17.087861 0.9940591
DunnettTest for selected Control Group
Dunnett’s test is kind of a multiple comparison procedure. Unlike Tukey’s test, it only compares your samples with the control group which could prevent the overestimation of the p-value because of non-related pairwise comparison.
|
Dunnett's test for comparing several treatments with a control : 95% family-wise confidence level $`1` diff lwr.ci upr.ci pval 2-1 30 13.58673988 46.41326 1.3e-05 *** 3-1 11 -3.04153165 25.04153 0.2062 4-1 29 14.54128620 43.45871 1.4e-06 *** 5-1 39 23.53916681 54.46083 8.7e-10 *** 6-1 17 2.95846835 31.04153 0.0091 ** 7-1 13 -0.02490811 26.02491 0.0505 . --- Signif. codes: 0 '\***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Two-way Anova
Reference: scribbr.com
When we needn’t to test any interaction between two groups, we could use the model W ~ C+G. Otherwise, W ~ C*G would be appropriate.
|
Call:
aov(formula = W ~ C * G, data = TB)
Terms:
C G C:G Residuals
Sum of Squares 57.1593 7.4347 420.1129 1479.5000
Deg. of Freedom 1 1 1 25
Residual standard error: 7.692854
Estimated effects may be unbalanced
|
Df Sum Sq Mean Sq F value Pr(>F) C 1 57.2 57.2 0.966 0.3351 G 1 7.4 7.4 0.126 0.7260 C:G 1 420.1 420.1 7.099 0.0133 * Residuals 25 1479.5 59.2 --- Signif. codes: 0 ‘\***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
According to the results from summary, neither Row (all men vs all women) nor column (all scandanavians vs all Americans) are significant. However, the interaction (C:G) p-value is 0.013, which is significant. This indicates that the relationships between ‘G’ and ‘W’ depends on the ‘C’ method.
codes from car:
Reference: R Statistics and Research, 2018
|
Anova Table (Type II tests)
Response: W
Sum Sq Df F value Pr(>F)
C 58.48 1 0.9881 0.32972
G 7.43 1 0.1256 0.72598
C:G 420.11 1 7.0989 0.01331 *
Residuals 1479.50 25
---
Signif. codes: 0 ‘\***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Repeat ANOVA measurement
You should different the repeated measure from other just like paired t test.
Reference: Repeated Measures ANOVA in R
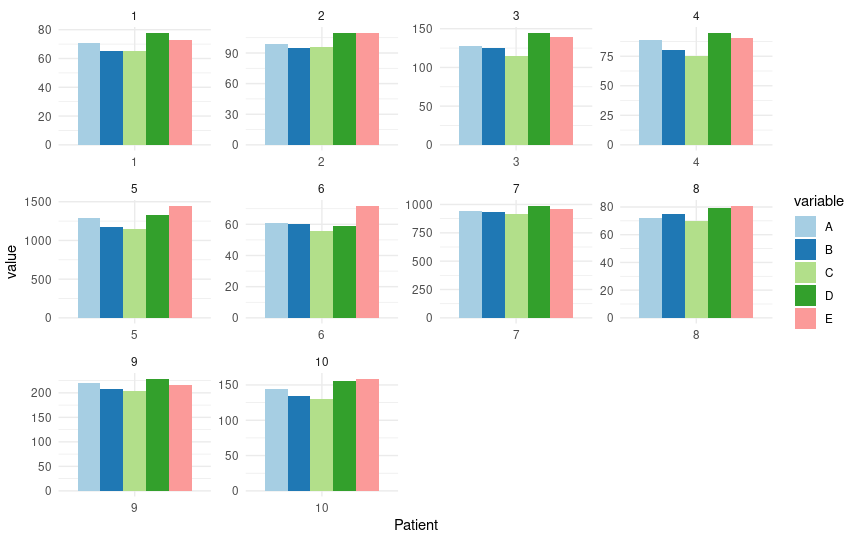
Ten patients did the same test from 5 different companies.
Save the data below as “123.txt”.
Patient A B C D E 1 71 65 65 78 73 2 99 95 96 110 110 3 128 125 115 145 139 4 89 80 75 95 90 5 1290 1180 1145 1330 1450 6 61 60 56 59 72 7 945 935 918 990 956 8 72 75 70 79 81 9 220 208 204 229 217 10 145 134 130 155 159
|
Patient A B C D E 1 1 71 65 65 78 73 2 2 99 95 96 110 110 3 3 128 125 115 145 139 4 4 89 80 75 95 90 5 5 1290 1180 1145 1330 1450 6 6 61 60 56 59 72
|
 |
|---|
With normal One-way anova:
|
Df Sum Sq Mean Sq F value Pr(>F) variable 4 16091 4023 0.021 0.999 Residuals 45 8465019 188112 Tukey multiple comparisons of means 95% family-wise confidence level Fit: aov(formula = value ~ variable, data = TB_g) $variable diff lwr upr p adj B-A -16.3 -567.441 534.841 0.9999882 C-A -24.6 -575.741 526.541 0.9999393 D-A 15.0 -536.141 566.141 0.9999916 E-A 22.7 -528.441 573.841 0.9999559 C-B -8.3 -559.441 542.841 0.9999992 D-B 31.3 -519.841 582.441 0.9998416 E-B 39.0 -512.141 590.141 0.9996211 D-C 39.6 -511.541 590.741 0.9995975 E-C 47.3 -503.841 598.441 0.9991880 E-D 7.7 -543.441 558.841 0.9999994
P value is very close to 1 which means that there are no significant difference between 5 companies.
This is worng because it treated all data as independent rather than repeated data
There is how we should deal with the repeated data:
|
ANOVA Table (type III tests)
$ANOVA
Effect DFn DFd F p p<.05 ges
1 variable 4 36 2.957 0.033 * 0.002
$`Mauchly's Test for Sphericity`
Effect W p p<.05
1 variable 8.46e-06 2.84e-14 *
$`Sphericity Corrections`
Effect GGe DF[GG] p[GG] p[GG]<.05 HFe DF[HF] p[HF] p[HF]<.05
1 variable 0.265 1.06, 9.55 0.117 0.271 1.08, 9.76 0.115
As you can see, the p value from anova result is 0.033 which is signifcant. On the other hand, the reuslt from “Mauchly’s test for Sphericity” is extremely signifcant. But After correction, the p-adjust is not significant. = =
For this data, we using Friedman Test.
The Friedman Test is a non-parametric alternative to the Repeated Measures ANOVA. It is used to determine whether or not there is a statistically significant difference between the means of three or more groups in which the same subjects show up in each group.
Zach from statology; 2020
|
# A tibble: 10 × 9 .y. group1 group2 n1 n2 statistic p p.adj p.adj.signif *1 value A B 10 10 52.5 0.012 0.125 ns 2 value A C 10 10 55 0.002 0.02 * 3 value A D 10 10 1 0.008 0.08 ns 4 value A E 10 10 3 0.014 0.138 ns 5 value B C 10 10 44 0.012 0.125 ns 6 value B D 10 10 1 0.008 0.08 ns 7 value B E 10 10 0 0.002 0.02 * 8 value C D 10 10 0 0.006 0.059 ns 9 value C E 10 10 0 0.002 0.02 * 10 value D E 10 10 26 0.722 1 ns
From the Friedman Test we can see 3 groups are significantly different.
To be notice that this result is slightly different from Prism for some reson. But it is the closed one to it after you choosed “Non parematic and Repeated” for doing ANOVA test.
Anova with rstatix library
Reference: rdrr.io
|
Chick Diet Wieght 1 : 1 1:16 Min. : 32.0 2 : 1 2:10 1st Qu.:126.0 3 : 1 3:10 Median :164.0 4 : 1 4: 9 Mean :177.6 5 : 1 3rd Qu.:225.0 6 : 1 Max. :332.0 (Other):39
|
Effect DFn DFd F p p<.05 ges 1 Diet 3 41 4.698 0.007 * 0.256
|
How to do anova in R