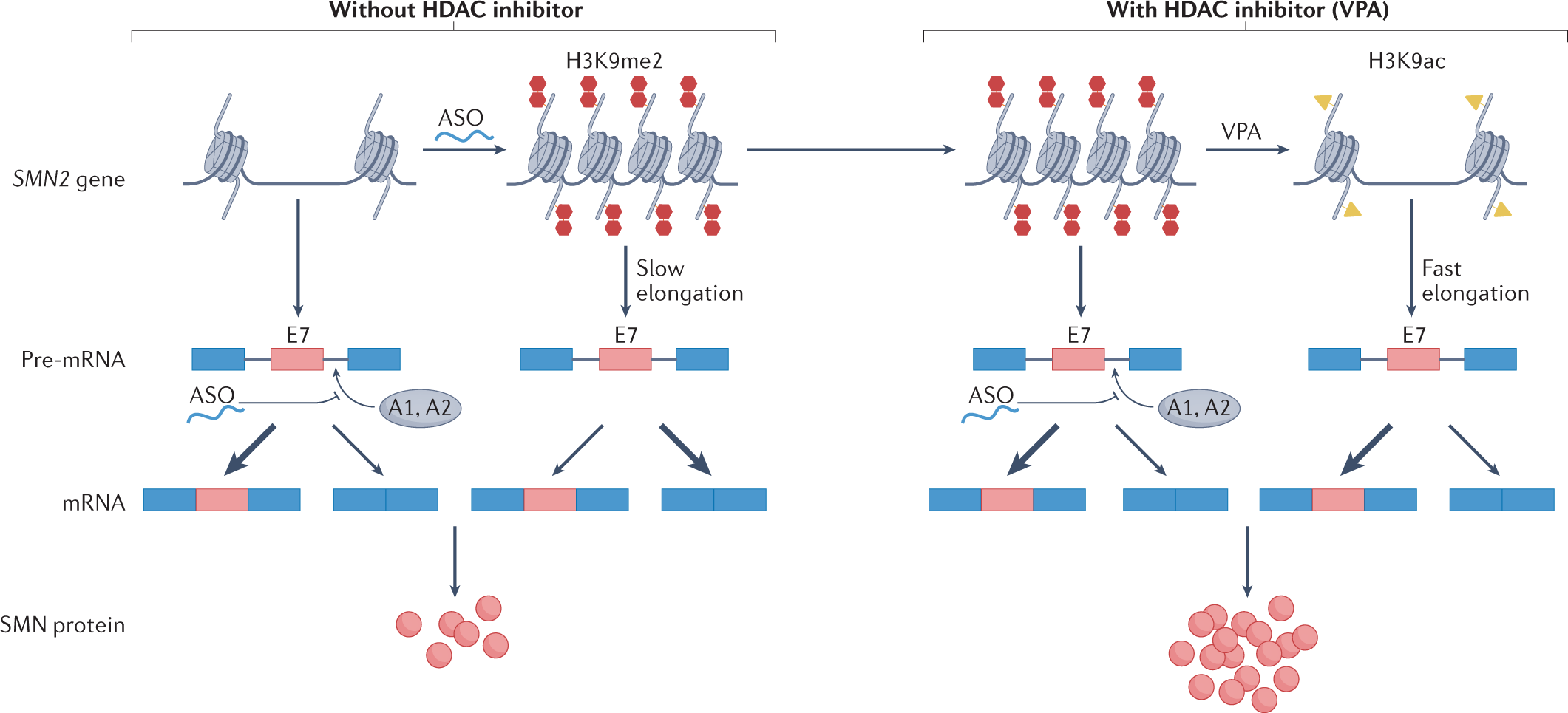
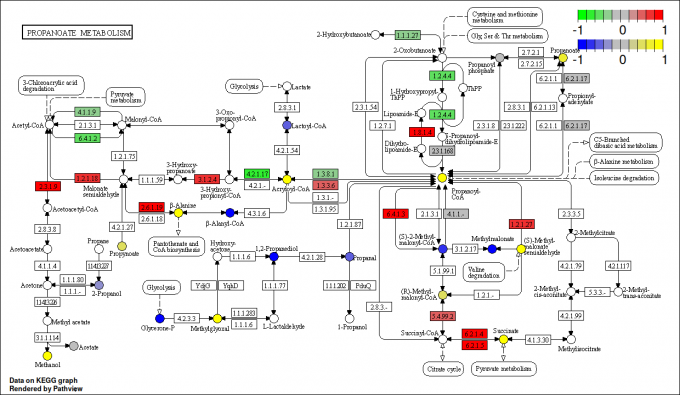
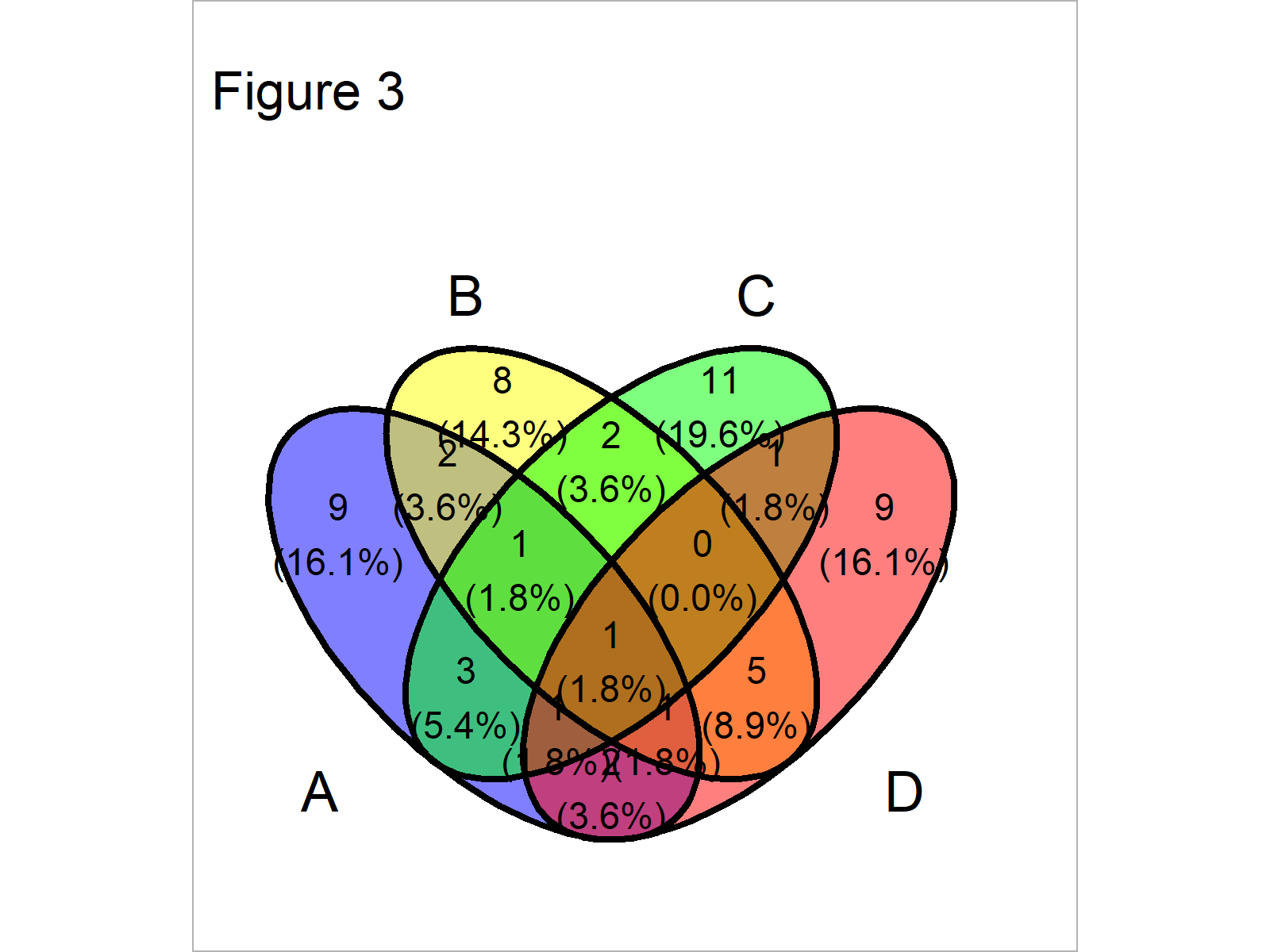
RNA Seq: Alternative Splicing
Alternative splicing research is important for understanding the diversity of gene expression and regulation, as it enables the production of multiple protein isoforms from a single gene, allowing cells to generate complex functional diversity and adapt to changing environmental conditions. Who said this?
Read more