Cell Ranger for scRNA-Seq Data Analysis
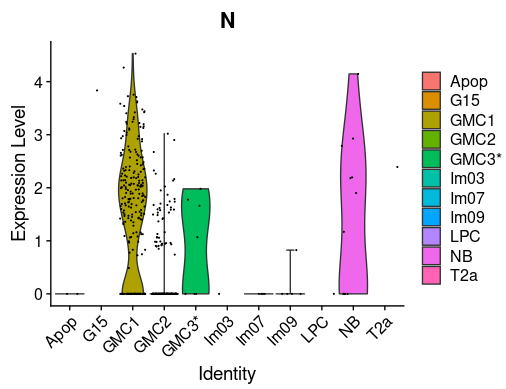
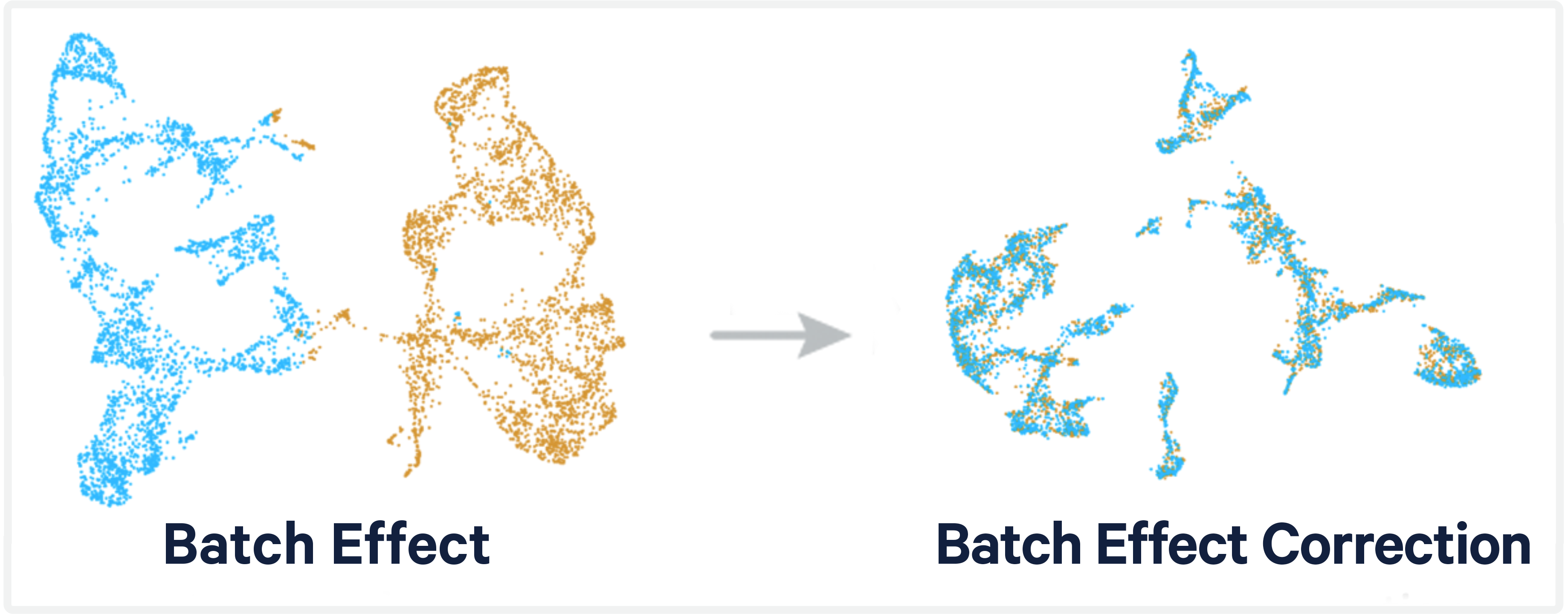
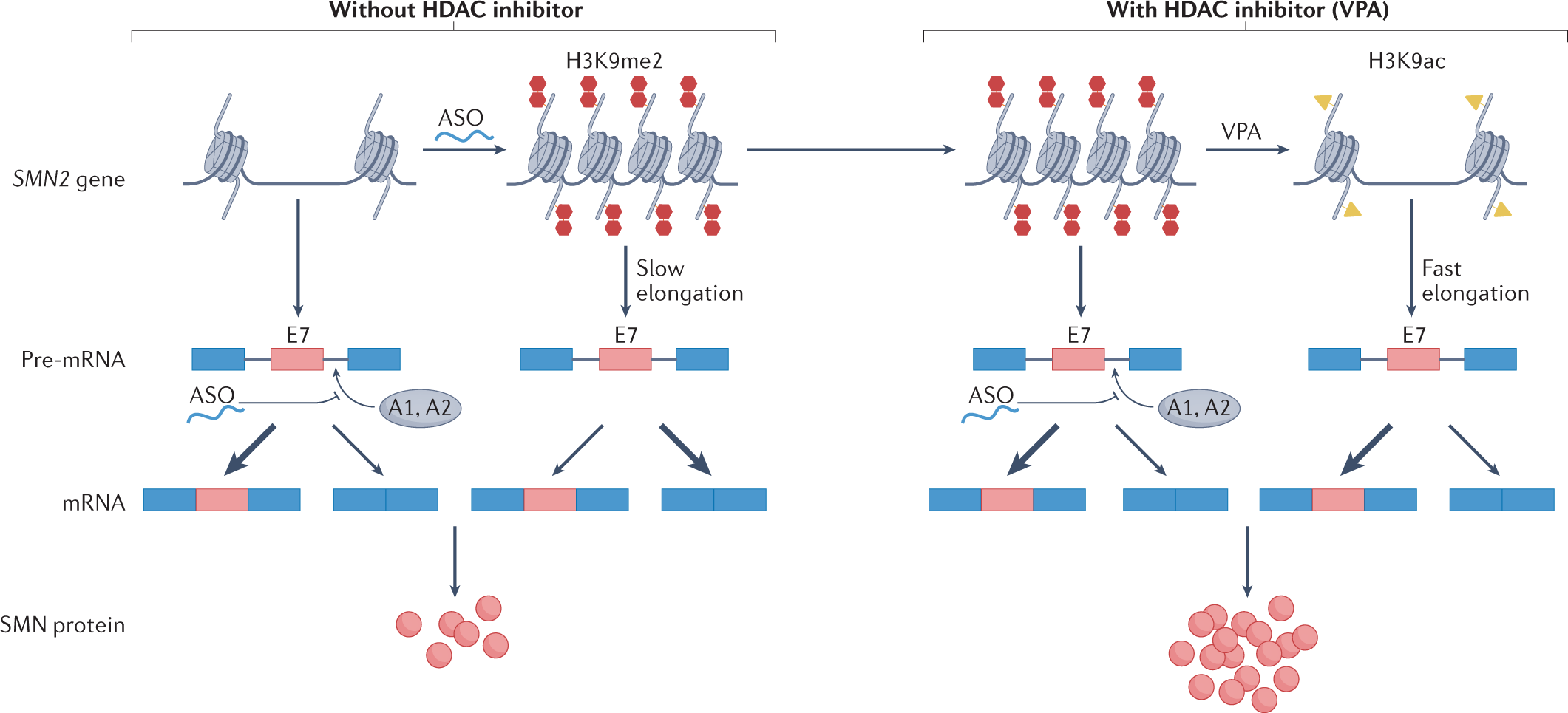
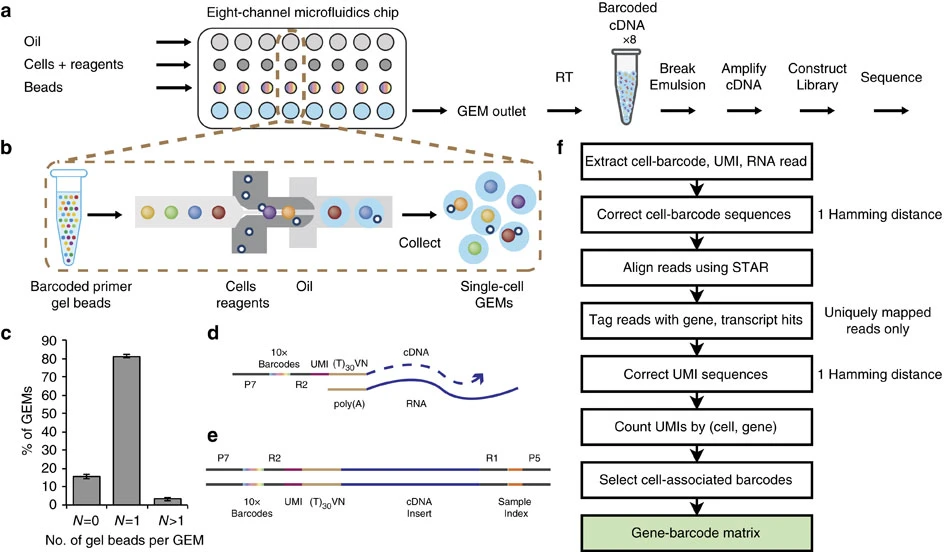
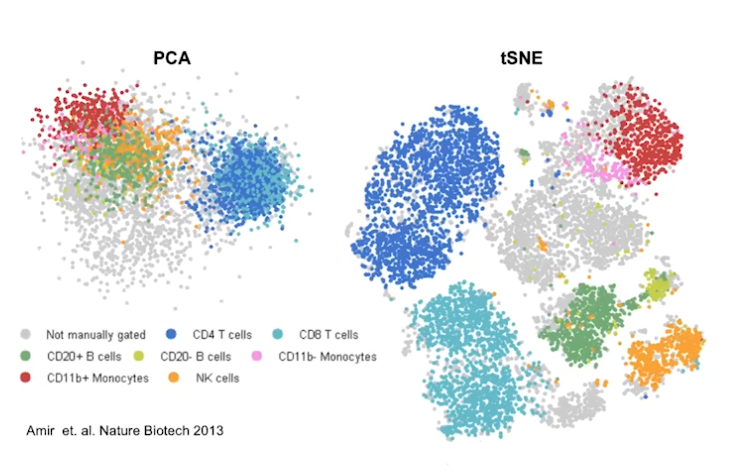
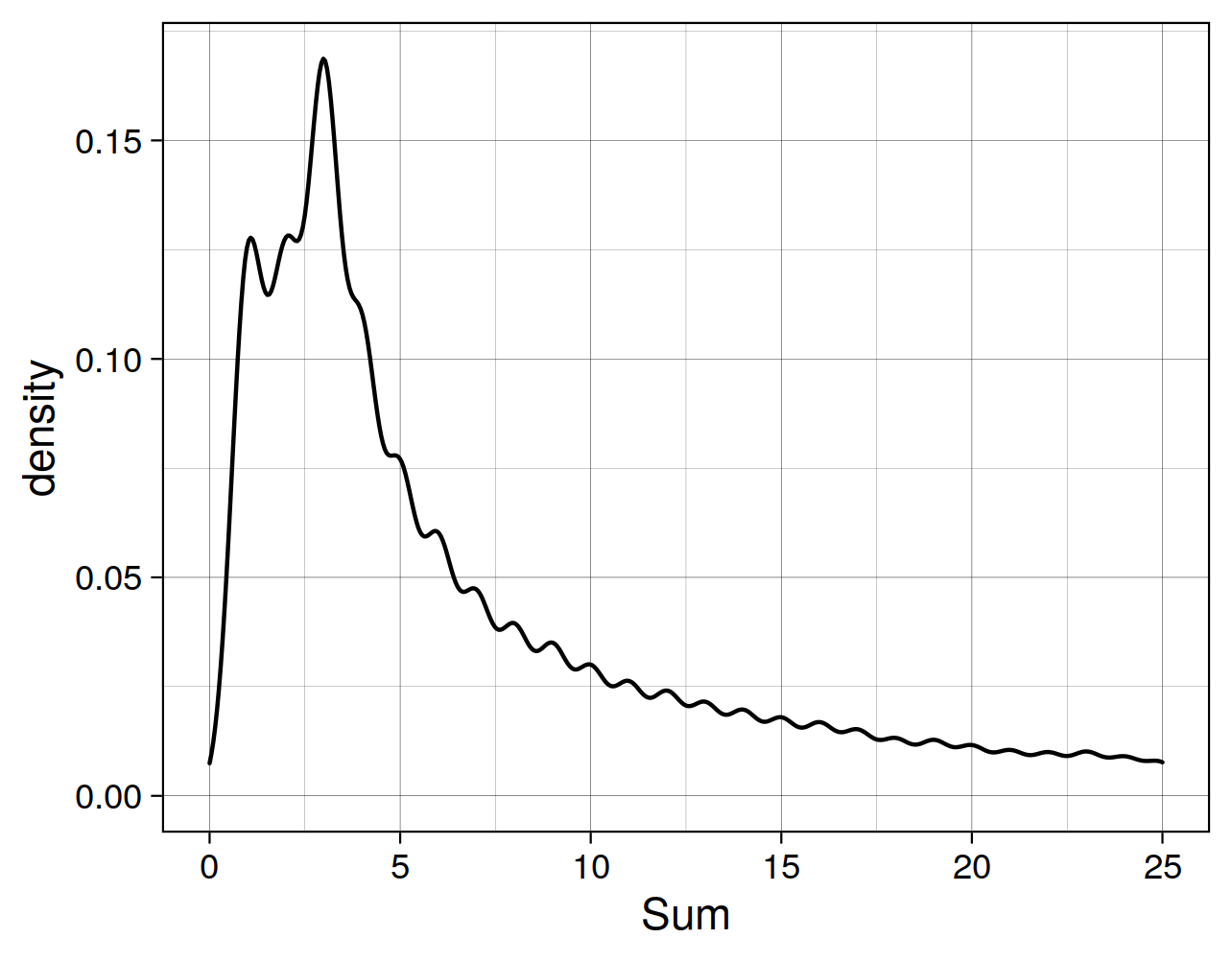
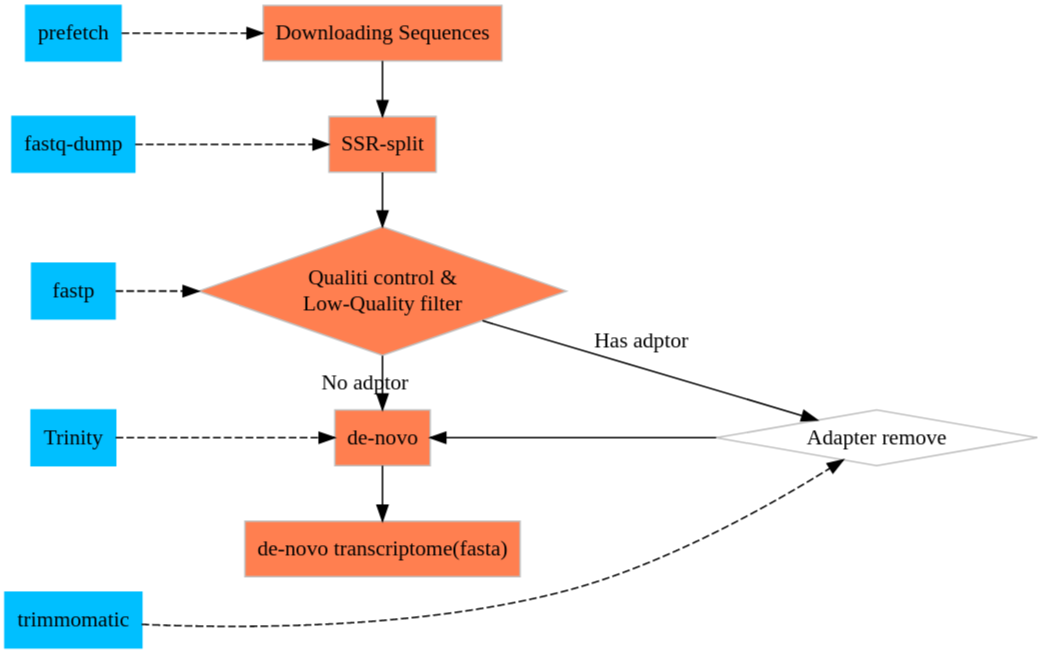
Cell Ranger is a software suite for analyzing single-cell RNA sequencing (scRNA-Seq) data, developed by 10x Genomics. It provides tools for processing raw sequencing data, performing quality control, and generating gene expression matrices.
Read more