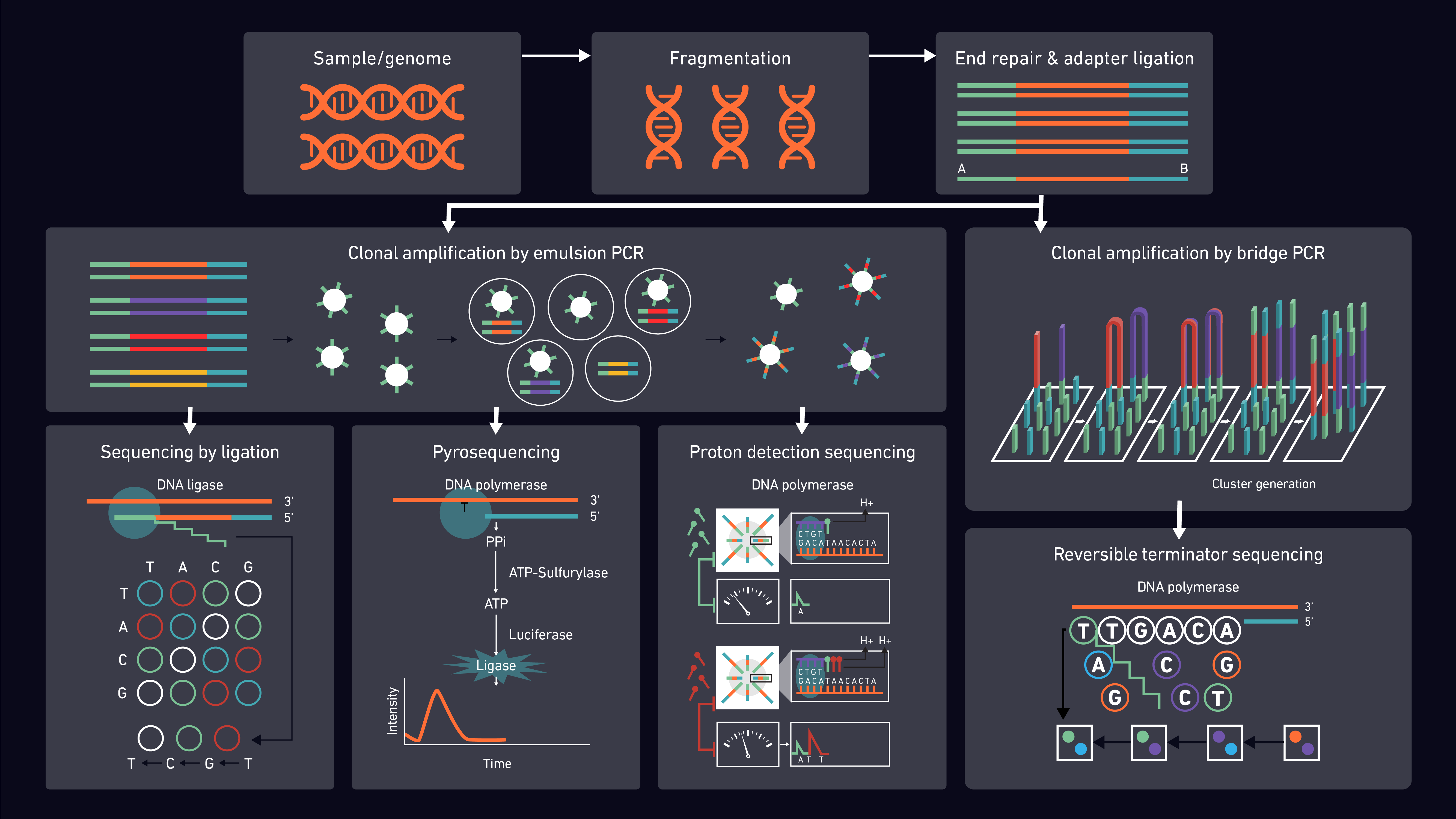
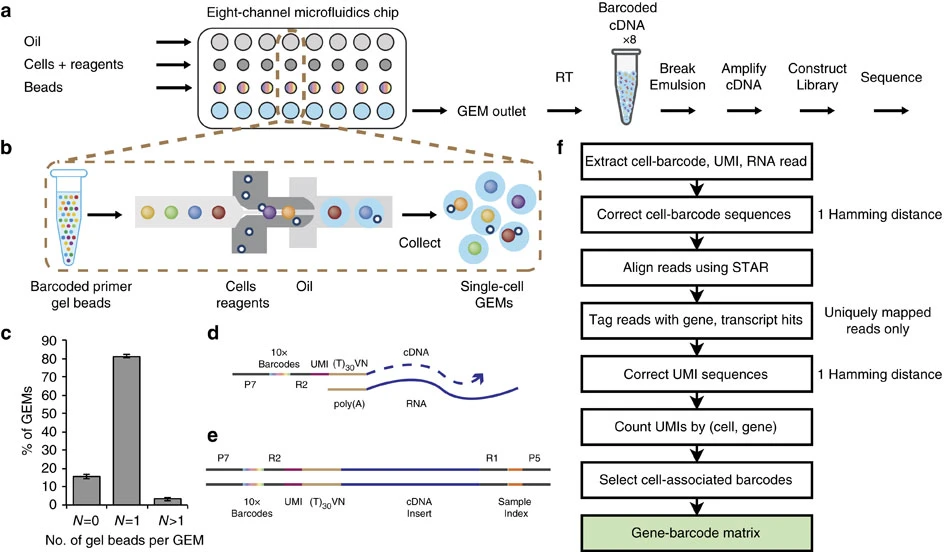
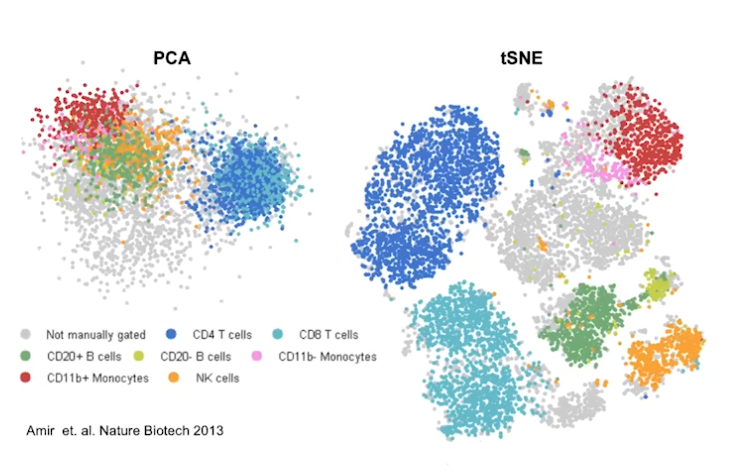
Short reads aligner compartment
The choice of software for aligning short reads in NGS can have a significant impact on the results. Different software programs use different algorithms for aligning reads and handling mismatches, leading to different levels of accuracy and sensitivity. Some software may prioritize speed, while others may prioritize accuracy, and still others may have specific strengths or limitations in handling certain types of data. It's important to carefully consider the characteristics and limitations of each software before making a choice to ensure the best possible alignment results Who sad this?
Read more